Computer-Assisted Processing of Current Step Signals in Single Blocking Impact Electrochemistry
IF 4.6
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
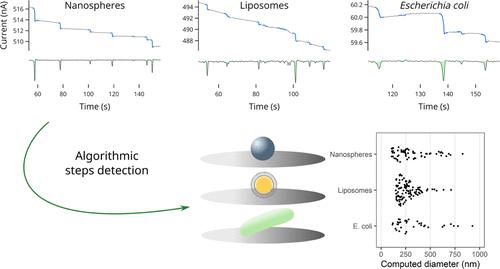
Current step signals related to single-entity collisions in blocking impact electrochemistry were analyzed by computer-assisted processing for estimating the size distributions of various particles. In this work, three different types of entities were studied by single blocking impact electrochemistry: polystyrene nanospheres (350 nm diameter) and microspheres (1 μm diameter), phospholipid liposomes (300 nm diameter) and two different strains of Gram-negative bacillus bacteria (Escherichia coli and Shewanella oneidensis). The size estimations of these different entities from the current step signal analysis were compared and discussed according to the shape and size of each entity. From the magnitude of the current step transient, the size distribution of each entity was calculated by a new computer program assisting in the detection and analysis of single impact events in chronoamperometry measurements. The data processing showed that the size distributions obtained from the electrochemical data agreed with the dynamic light scattering and atomic force microscopy data for nanospheres and liposomes. In contrast, the size estimation calculated from the electrochemical data was underestimated for microspheres and bacteria. We demonstrated that our computer program was efficient for detecting and analyzing the collision events in single blocking impact electrochemistry for various entities from spherical hard nanoparticles to micrometer-sized rod-shaped living bacteria.
计算机辅助处理单阻断影响电化学中的电流阶跃信号
通过计算机辅助处理,分析了阻断冲击电化学中与单实体碰撞有关的电流阶跃信号,以估计各种粒子的尺寸分布。在这项工作中,通过单次阻塞撞击电化学研究了三种不同类型的实体:聚苯乙烯纳米球(直径为 350 nm)和微球(直径为 1 μm)、磷脂脂质体(直径为 300 nm)以及两种不同的革兰氏阴性杆菌菌株(大肠杆菌和 Shewanella oneidensis)。根据每个实体的形状和大小,对电流阶跃信号分析得出的这些不同实体的大小估计进行了比较和讨论。根据电流阶跃瞬态的大小,用一个新的计算机程序计算出了每个实体的大小分布,该程序可协助检测和分析计时器振荡测量中的单次冲击事件。数据处理结果表明,从电化学数据中获得的尺寸分布与纳米球和脂质体的动态光散射和原子力显微镜数据一致。相反,对于微球和细菌,根据电化学数据计算出的粒度估计值被低估了。我们证明了我们的计算机程序可以有效地检测和分析从球形硬质纳米粒子到微米大小的杆状活细菌等各种实体在单阻挡撞击电化学中的碰撞事件。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Measurement Science Au
化学计量学-
CiteScore
5.20
自引率
0.00%
发文量
0
期刊介绍:
ACS Measurement Science Au is an open access journal that publishes experimental computational or theoretical research in all areas of chemical measurement science. Short letters comprehensive articles reviews and perspectives are welcome on topics that report on any phase of analytical operations including sampling measurement and data analysis. This includes:Chemical Reactions and SelectivityChemometrics and Data ProcessingElectrochemistryElemental and Molecular CharacterizationImagingInstrumentationMass SpectrometryMicroscale and Nanoscale systemsOmics (Genomics Proteomics Metabonomics Metabolomics and Bioinformatics)Sensors and Sensing (Biosensors Chemical Sensors Gas Sensors Intracellular Sensors Single-Molecule Sensors Cell Chips Arrays Microfluidic Devices)SeparationsSpectroscopySurface analysisPapers dealing with established methods need to offer a significantly improved original application of the method.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


