Integrating large-scale single-cell RNA sequencing in central nervous system disease using self-supervised contrastive learning
IF 5.2
1区 生物学
Q1 BIOLOGY
引用次数: 0
Abstract
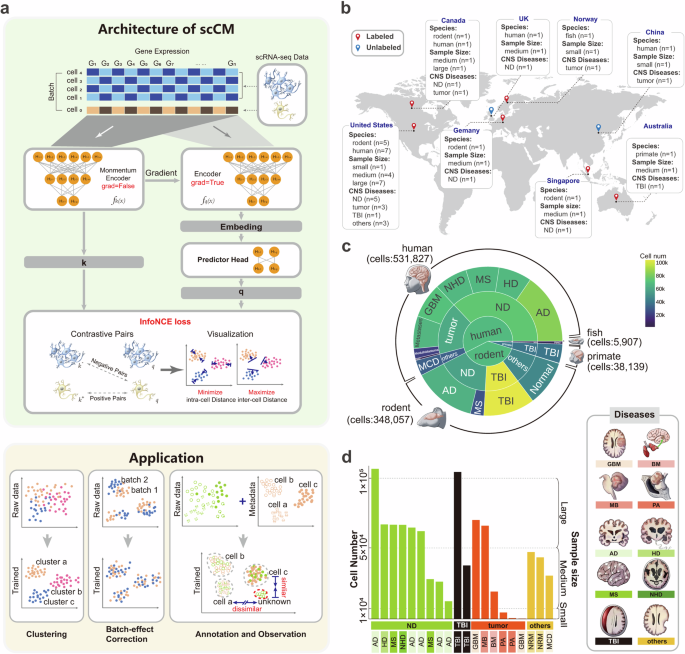
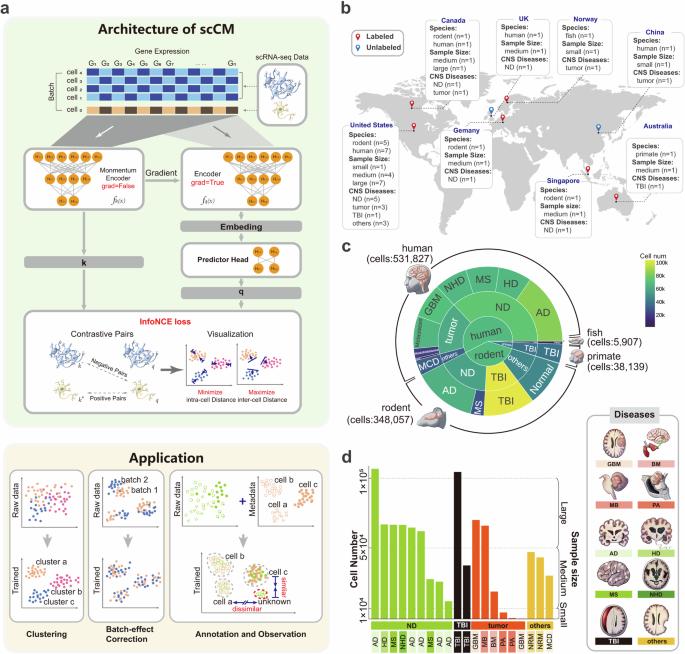
The central nervous system (CNS) comprises a diverse range of brain cell types with distinct functions and gene expression profiles. Although single-cell RNA sequencing (scRNA-seq) provides new insights into the brain cell atlases, integrating large-scale CNS scRNA-seq data still encounters challenges due to the complexity and heterogeneity among CNS cell types/subtypes. In this study, we introduce a self-supervised contrastive learning method, called scCM, for integrating large-scale CNS scRNA-seq data. scCM brings functionally related cells close together while simultaneously pushing apart dissimilar cells by comparing the variations of gene expression, effectively revealing the heterogeneous relationships within the CNS cell types/subtypes. The effectiveness of scCM is evaluated on 20 CNS datasets covering 4 species and 10 CNS diseases. Leveraging these strengths, we successfully integrate the collected human CNS datasets into a large-scale reference to annotate cell types and subtypes in neural tissues. Results demonstrate that scCM provides an accurate annotation, along with rich spatial information of cell state. In summary, scCM is a robust and promising method for integrating large-scale CNS scRNA-seq data, enabling researchers to gain insights into the cellular and molecular mechanisms underlying CNS functions and diseases. scCM, a self-supervised contrastive learning method, effectively integrates large-scale CNS scRNA-seq data by clustering functionally related cells and separating dissimilar ones.
利用自监督对比学习整合中枢神经系统疾病中的大规模单细胞 RNA 测序
中枢神经系统(CNS)由多种脑细胞类型组成,它们具有不同的功能和基因表达谱。虽然单细胞 RNA 测序(scRNA-seq)为脑细胞图谱提供了新的见解,但由于中枢神经系统细胞类型/亚型之间的复杂性和异质性,整合大规模中枢神经系统 scRNA-seq 数据仍面临挑战。在本研究中,我们介绍了一种用于整合大规模中枢神经系统 scRNA-seq 数据的自监督对比学习方法,称为 scCM。scCM 通过比较基因表达的变化,将功能相关的细胞靠近在一起,同时将不相似的细胞推开,有效揭示了中枢神经系统细胞类型/亚型之间的异质性关系。我们在涵盖 4 个物种和 10 种中枢神经系统疾病的 20 个中枢神经系统数据集上评估了 scCM 的有效性。利用这些优势,我们成功地将收集到的人类中枢神经系统数据集整合成一个大规模参考资料,用于注释神经组织中的细胞类型和亚型。结果表明,scCM 提供了准确的注释以及丰富的细胞状态空间信息。总之,scCM 是一种用于整合大规模中枢神经系统 scRNA-seq 数据的稳健而有前景的方法,能让研究人员深入了解中枢神经系统功能和疾病的细胞和分子机制。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Communications Biology
Medicine-Medicine (miscellaneous)
CiteScore
8.60
自引率
1.70%
发文量
1233
审稿时长
13 weeks
期刊介绍:
Communications Biology is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the biological sciences. Research papers published by the journal represent significant advances bringing new biological insight to a specialized area of research.
文献相关原料
| 公司名称 | 产品信息 | 采购帮参考价格 |
|---|
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


