Colorimetric Hybridization Sensor for DNA Mimic of a SARS-CoV-2 RNA Marker: Direct and Inverse Bioanalysis
IF 4.6
Q1 CHEMISTRY, ANALYTICAL
引用次数: 0
Abstract
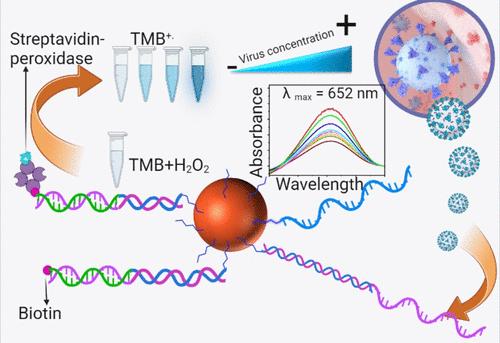
This article presents a colorimetric visual biosensor designed for direct application in undiluted biofluids, which holds significant promise for point-of-need applications. Unlike traditional biosensors that struggle with heavily diluted sample matrices, the presented biosensor does not require any instrumentation or trained personnel, making it highly practical. The sensor features an oligonucleotide probe covalently attached to magnetically separable magnetite (Fe3O4) particles. This probe selectively captures a DNA mimic of the SARS-CoV-2 RNA sequence via a base-pair hybridization. The DNA mimic oligomer sequence was tested in a buffer solution, undiluted serum, and undiluted salivary biofluids. A second complementary hybridization sequence with a biotin tag was used to bind the target oligomer already hybridized to the magnetic particle-conjugated capture probe. Subsequent detection of the target oligomer was accomplished through high-affinity selective binding of streptavidin-peroxidase labels with the detection probe biotin units for visual colorimetric detection in the presence of 3,3′,5,5′-tetramethylbenzidine and hydrogen peroxide. Inverse assaying of the unbound-free streptavidin-peroxidase labels left in the detection reagent solution offered a reverse trend to the target oligomer concentration, as anticipated. We obtained detection limits of 1 fM (buffer assay), 1 pM (undiluted serum assay), and 1 pM (undiluted saliva assay) and with the linear ranges of 1 fM–10 nM (buffer assay), 1 pM–1 nM (undiluted serum assay), and 1 pM–1 nM (undiluted saliva assay), respectively. The assays in different biofluids allowed for the estimation of the analytical performance and the effect of sample matrices on the detection limits and calibration sensitivity.
用于 SARS-CoV-2 RNA 标记 DNA 模拟的比色杂交传感器:直接和逆向生物分析
本文介绍了一种可直接应用于未稀释生物流体的比色视觉生物传感器,它在需求点应用方面前景广阔。传统的生物传感器在严重稀释的样品基质中难以发挥作用,而本文介绍的生物传感器则不同,它不需要任何仪器或训练有素的人员,因此非常实用。该传感器将寡核苷酸探针共价连接到磁性可分离的磁铁矿(Fe3O4)颗粒上。该探针通过碱基对杂交选择性地捕获 SARS-CoV-2 RNA 序列的 DNA 模拟物。在缓冲溶液、未稀释的血清和未稀释的唾液生物流体中测试了 DNA 模拟寡聚体序列。第二个带有生物素标记的互补杂交序列用于结合已经与磁性颗粒结合的捕获探针杂交的目标寡聚体。在 3,3′,5,5′-四甲基联苯胺和过氧化氢存在的情况下,通过链霉亲和素-过氧化物酶标签与检测探针生物素单位的高亲和性选择性结合进行可视比色检测,从而实现对目标寡聚体的后续检测。对检测试剂溶液中残留的未结合的链霉亲和素-过氧化物酶标签进行反向测定,结果与预期的目标低聚物浓度呈反向趋势。我们得到的检测限分别为 1 fM(缓冲液检测)、1 pM(未稀释血清检测)和 1 pM(未稀释唾液检测),线性范围分别为 1 fM-10 nM(缓冲液检测)、1 pM-1 nM(未稀释血清检测)和 1 pM-1 nM(未稀释唾液检测)。在不同的生物流体中进行测定,可以评估分析性能以及样品基质对检测限和校准灵敏度的影响。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Measurement Science Au
化学计量学-
CiteScore
5.20
自引率
0.00%
发文量
0
期刊介绍:
ACS Measurement Science Au is an open access journal that publishes experimental computational or theoretical research in all areas of chemical measurement science. Short letters comprehensive articles reviews and perspectives are welcome on topics that report on any phase of analytical operations including sampling measurement and data analysis. This includes:Chemical Reactions and SelectivityChemometrics and Data ProcessingElectrochemistryElemental and Molecular CharacterizationImagingInstrumentationMass SpectrometryMicroscale and Nanoscale systemsOmics (Genomics Proteomics Metabonomics Metabolomics and Bioinformatics)Sensors and Sensing (Biosensors Chemical Sensors Gas Sensors Intracellular Sensors Single-Molecule Sensors Cell Chips Arrays Microfluidic Devices)SeparationsSpectroscopySurface analysisPapers dealing with established methods need to offer a significantly improved original application of the method.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


