Analysis of Complexome Profiles with the Gaussian Interaction Profiler (GIP) Reveals Novel Protein Complexes in Plasmodium falciparum
IF 4.3
3区 材料科学
Q1 ENGINEERING, ELECTRICAL & ELECTRONIC
引用次数: 0
Abstract
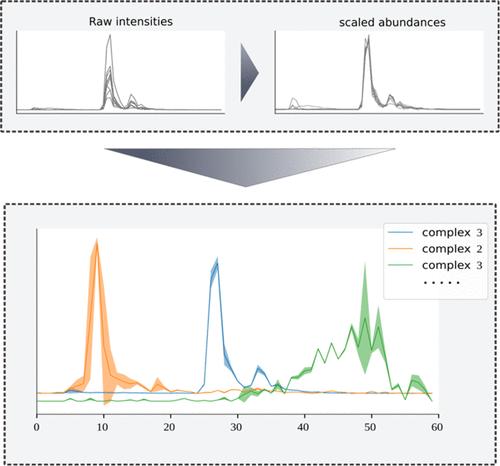
Complexome profiling is an experimental approach to identify interactions by integrating native separation of protein complexes and quantitative mass spectrometry. In a typical complexome profile, thousands of proteins are detected across typically ≤100 fractions. This relatively low resolution leads to similar abundance profiles between proteins that are not necessarily interaction partners. To address this challenge, we introduce the Gaussian Interaction Profiler (GIP), a Gaussian mixture modeling-based clustering workflow that assigns protein clusters by modeling the migration profile of each cluster. Uniquely, the GIP offers a way to prioritize actual interactors over spuriously comigrating proteins. Using previously analyzed human fibroblast complexome profiles, we show good performance of the GIP compared to other state-of-the-art tools. We further demonstrate GIP utility by applying it to complexome profiles from the transmissible lifecycle stage of malaria parasites. We unveil promising novel associations for future experimental verification, including an interaction between the vaccine target Pfs47 and the hypothetical protein PF3D7_0417000. Taken together, the GIP provides methodological advances that facilitate more accurate and automated detection of protein complexes, setting the stage for more varied and nuanced analyses in the field of complexome profiling. The complexome profiling data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD050751.
用高斯相互作用剖析器(GIP)分析复合物组图谱,揭示恶性疟原虫中的新型蛋白质复合物
复合体组剖析是一种通过整合蛋白质复合体的原生分离和定量质谱法来识别相互作用的实验方法。在一个典型的复合物组图谱中,通常≤100 个馏分可检测到数千个蛋白质。这种相对较低的分辨率会导致不一定是相互作用伙伴的蛋白质之间出现相似的丰度曲线。为了应对这一挑战,我们引入了高斯相互作用剖析器(GIP),这是一种基于高斯混合建模的聚类工作流程,通过对每个聚类的迁移谱建模来分配蛋白质聚类。与众不同的是,GIP 提供了一种方法来优先考虑实际的相互作用者,而不是虚假的合并蛋白。利用之前分析的人类成纤维细胞复合体图谱,我们展示了 GIP 与其他最先进工具相比的良好性能。通过将 GIP 应用于疟疾寄生虫可传播生命周期阶段的复合体图谱,我们进一步证明了 GIP 的实用性。我们揭示了有希望在未来进行实验验证的新关联,包括疫苗靶标 Pfs47 与假想蛋白 PF3D7_0417000 之间的相互作用。总之,GIP 在方法学方面取得了进步,有助于更准确、更自动化地检测蛋白质复合物,为在复合物组剖析领域进行更多样、更细致的分析奠定了基础。复合物组分析数据已存入蛋白质组交换联盟(ProteomeXchange Consortium),数据集标识符为 PXD050751。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


