Development and Validation of a Predictive Model for Resistance to Platinum-Based Chemotherapy in Patients with Ovarian Cancer through Proteomic Analysis
IF 3.6
2区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Platinum resistance in ovarian cancer poses a significant challenge, substantially impacting patient outcomes. Developing an accurate predictive model is crucial for improving clinical decision-making and guiding treatment strategies. Proteomic data from 217 high-grade serous ovarian cancer (HGSOC) biospecimens obtained from JHU, PNNL, and PTRC were used to construct a prediction model for identifying individuals who are resistant to platinum-based chemotherapy. A total of 6437 common proteins were detected across all data sets, with 26 proteins overlapping between the development cohorts JHU and PNNL. Using LASSO and logistic regression analysis, a six-protein model (P31323_PRKAR2B, Q13309_SKP2, Q14997_PSME4, Q6ZRP7_QSOX2, Q7LGA3_HS2ST1, and Q7Z2Z2_EFL1) was developed, which accurately predicted platinum resistance, with an AUC of 0.964 (95% CI, 0.929–0.999). Internal validation by resampling resulted in a C-index of 0.972 (95% CI 0.894–0.988). External validation performed on the PTRC cohort achieved an AUC of 0.855 (95% CI 0.748–0.963). Calibration curves showed good consistency, and DCA indicated superior clinical utility. The model also performed well in predicting PFS and OS at various time points. Based on these proteins, our predictive model can precisely predict platinum response and survival outcomes in HGSOC patients, which can assist clinicians in promptly identifying potentially platinum-resistant individuals.
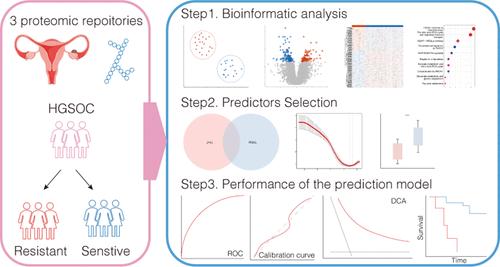
通过蛋白质组分析建立并验证卵巢癌患者对铂类化疗耐药性的预测模型
卵巢癌的铂耐药性是一项重大挑战,严重影响患者的预后。建立准确的预测模型对于改善临床决策和指导治疗策略至关重要。我们利用从哈佛大学、PNNL和PTRC获得的217份高级别浆液性卵巢癌(HGSOC)生物样本的蛋白质组数据构建了一个预测模型,用于识别对铂类化疗耐药的个体。所有数据集共检测到 6437 个常见蛋白质,其中有 26 个蛋白质在 JHU 和 PNNL 研发队列中重叠。利用 LASSO 和逻辑回归分析,建立了一个六种蛋白质模型(P31323_PRKAR2B、Q13309_SKP2、Q14997_PSME4、Q6ZRP7_QSOX2、Q7LGA3_HS2ST1 和 Q7Z2Z2_EFL1),该模型能准确预测铂金耐药性,AUC 为 0.964(95% CI,0.929-0.999)。通过重采样进行内部验证后,C 指数为 0.972(95% CI 0.894-0.988)。在 PTRC 队列中进行的外部验证的 AUC 为 0.855(95% CI 0.748-0.963)。校准曲线显示出良好的一致性,DCA 显示出卓越的临床实用性。该模型在预测不同时间点的 PFS 和 OS 方面也表现良好。基于这些蛋白质,我们的预测模型可以精确预测HGSOC患者的铂反应和生存结果,从而帮助临床医生及时发现潜在的铂耐药个体。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Proteome Research
生物-生化研究方法
CiteScore
9.00
自引率
4.50%
发文量
251
审稿时长
3 months
期刊介绍:
Journal of Proteome Research publishes content encompassing all aspects of global protein analysis and function, including the dynamic aspects of genomics, spatio-temporal proteomics, metabonomics and metabolomics, clinical and agricultural proteomics, as well as advances in methodology including bioinformatics. The theme and emphasis is on a multidisciplinary approach to the life sciences through the synergy between the different types of "omics".
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


