Disease prediction with multi-omics and biomarkers empowers case–control genetic discoveries in the UK Biobank
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
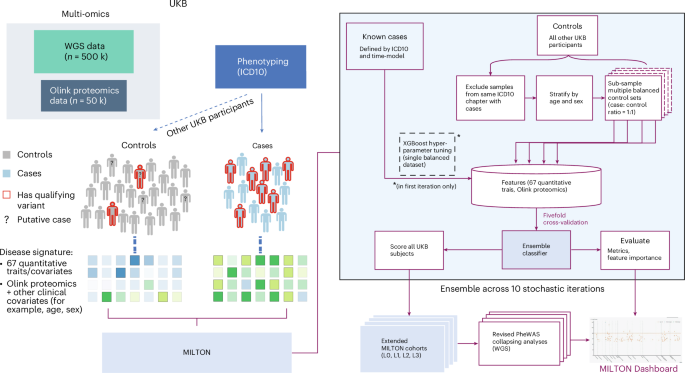
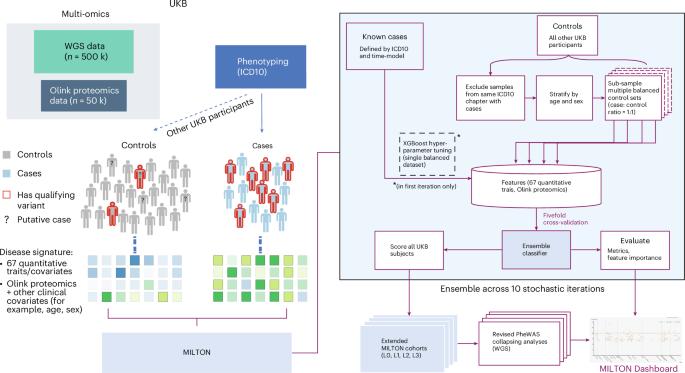
The emergence of biobank-level datasets offers new opportunities to discover novel biomarkers and develop predictive algorithms for human disease. Here, we present an ensemble machine-learning framework (machine learning with phenotype associations, MILTON) utilizing a range of biomarkers to predict 3,213 diseases in the UK Biobank. Leveraging the UK Biobank’s longitudinal health record data, MILTON predicts incident disease cases undiagnosed at time of recruitment, largely outperforming available polygenic risk scores. We further demonstrate the utility of MILTON in augmenting genetic association analyses in a phenome-wide association study of 484,230 genome-sequenced samples, along with 46,327 samples with matched plasma proteomics data. This resulted in improved signals for 88 known (P < 1 × 10−8) gene–disease relationships alongside 182 gene–disease relationships that did not achieve genome-wide significance in the nonaugmented baseline cohorts. We validated these discoveries in the FinnGen biobank alongside two orthogonal machine-learning methods built for gene–disease prioritization. All extracted gene–disease associations and incident disease predictive biomarkers are publicly available ( http://milton.public.cgr.astrazeneca.com ). MILTON uses phenotype information in the UK Biobank to identify clinical biomarkers and other quantitative traits that characterize diseases. It then constructs augmented cohorts by predicting undiagnosed individuals, improving power to discover gene–disease relationships.
利用多组学和生物标志物进行疾病预测有助于英国生物库中的病例对照基因发现
生物库级数据集的出现为发现新型生物标记物和开发人类疾病预测算法提供了新的机遇。在这里,我们提出了一个集合机器学习框架(表型关联机器学习,MILTON),利用一系列生物标记物预测英国生物库中的 3213 种疾病。利用英国生物库的纵向健康记录数据,MILTON 可以预测招募时未确诊的疾病病例,在很大程度上优于现有的多基因风险评分。我们在一项全表型关联研究中进一步证明了 MILTON 在增强遗传关联分析方面的实用性,该研究包括 484,230 份基因组测序样本以及 46,327 份具有匹配血浆蛋白质组学数据的样本。这使得 88 种已知(P < 1 × 10-8)基因-疾病关系以及 182 种在非增强基线队列中未达到全基因组显著性的基因-疾病关系的信号得到改善。我们在 FinnGen 生物库中用两种正交的机器学习方法验证了这些发现,这两种方法都是为基因疾病优先排序而建立的。所有提取的基因-疾病关联和事件疾病预测生物标记物均可公开获取 (http://milton.public.cgr.astrazeneca.com)。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


