Gut microbial subtypes and clinicopathological value for colorectal cancer
Abstract
Background
Gut bacteria are related to colorectal cancer (CRC) and its clinicopathologic characteristics.
Objective
To develop gut bacterial subtypes and explore potential microbial targets for CRC.
Methods
Stool samples from 914 volunteers (376 CRCs, 363 advanced adenomas, and 175 normal controls) were included for 16S rRNA sequencing. Unsupervised learning was used to generate gut microbial subtypes. Gut bacterial community composition and clustering effects were plotted. Differences of gut bacterial abundance were analyzed. Then, the association of CRC-associated bacteria with subtypes and the association of gut bacteria with clinical information were assessed. The CatBoost models based on gut differential bacteria were constructed to identify the diseases including CRC and advanced adenoma (AA).
Results
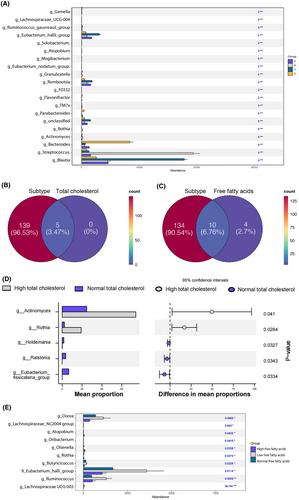
Four gut microbial subtypes (A, B, C, D) were finally obtained via unsupervised learning. The characteristic bacteria of each subtype were Escherichia-Shigella in subtype A, Streptococcus in subtype B, Blautia in subtype C, and Bacteroides in subtype D. Clinical information (e.g., free fatty acids and total cholesterol) and CRC pathological information (e.g., tumor depth) varied among gut microbial subtypes. Bacilli, Lactobacillales, etc., were positively correlated with subtype B. Positive correlation of Blautia, Lachnospiraceae, etc., with subtype C and negative correlation of Coriobacteriia, Coriobacteriales, etc., with subtype D were found. Finally, the predictive ability of CatBoost models for CRC identification was improved based on gut microbial subtypes.
Conclusion
Gut microbial subtypes provide characteristic gut bacteria and are expected to contribute to the diagnosis of CRC.
| 公司名称 | 产品信息 | 采购帮参考价格 |
|---|
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


