Single-cell detection of copy number changes reveals dynamic mechanisms of adaptation to antifungals in Candida albicans
IF 20.5
1区 生物学
Q1 MICROBIOLOGY
引用次数: 0
Abstract
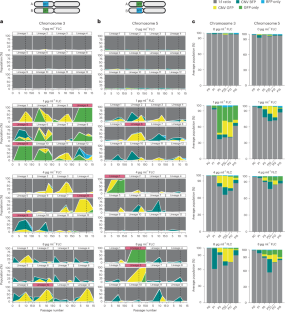
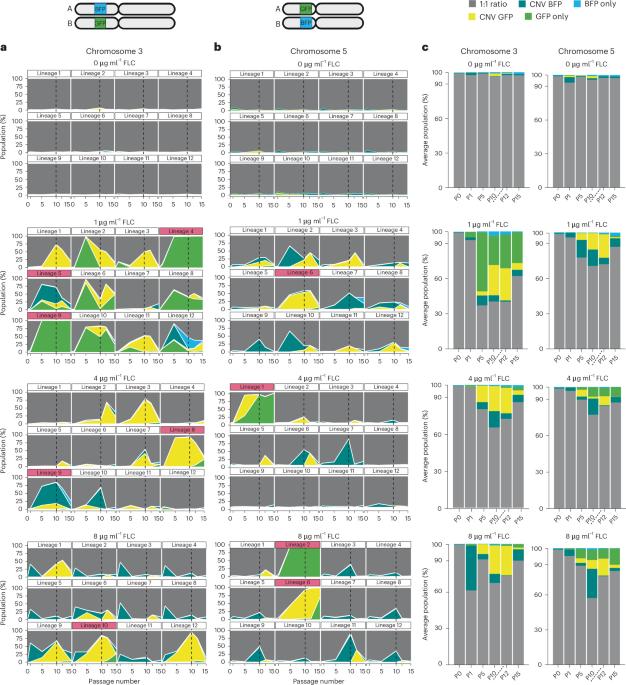
Genomic copy number changes are associated with antifungal drug resistance and virulence across diverse fungal pathogens, but the rate and dynamics of these genomic changes in the presence of antifungal drugs are unknown. Here we optimized a dual-fluorescent reporter system in the diploid pathogen Candida albicans to quantify haplotype-specific copy number variation (CNV) and loss of heterozygosity (LOH) at the single-cell level with flow cytometry. We followed the frequency and dynamics of CNV and LOH at two distinct genomic locations in the presence and absence of antifungal drugs in vitro and in a murine model of candidiasis. Copy number changes were rapid and dynamic during adaptation to fluconazole and frequently involved competing subpopulations with distinct genotypes. This study provides quantitative evidence for the rapid speed at which diverse genotypes arise and undergo dynamic population-level fluctuations during adaptation to antifungal drugs in vitro and in vivo. Development of a dual-fluorescent reporter system allows rapid quantification of diverse genomic copy number changes that arise in Candida albicans during adaptation to antifungal drugs.
单细胞检测拷贝数变化揭示白念珠菌对抗真菌药物的动态适应机制
基因组拷贝数变化与各种真菌病原体的抗真菌药物耐药性和毒力有关,但这些基因组变化在抗真菌药物作用下的速率和动态尚不清楚。在这里,我们优化了二倍体病原体白色念珠菌的双荧光报告系统,利用流式细胞仪在单细胞水平上量化单倍型特异性拷贝数变异(CNV)和杂合性缺失(LOH)。我们跟踪了体外和小鼠念珠菌病模型中抗真菌药物存在和不存在时,两个不同基因组位置的 CNV 和 LOH 的频率和动态变化。在对氟康唑的适应过程中,拷贝数的变化是快速和动态的,而且经常涉及具有不同基因型的竞争亚群。这项研究提供了定量证据,证明在体外和体内适应抗真菌药物的过程中,不同基因型的出现和种群水平的动态波动速度很快。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Microbiology
Immunology and Microbiology-Microbiology
CiteScore
44.40
自引率
1.10%
发文量
226
期刊介绍:
Nature Microbiology aims to cover a comprehensive range of topics related to microorganisms. This includes:
Evolution: The journal is interested in exploring the evolutionary aspects of microorganisms. This may include research on their genetic diversity, adaptation, and speciation over time.
Physiology and cell biology: Nature Microbiology seeks to understand the functions and characteristics of microorganisms at the cellular and physiological levels. This may involve studying their metabolism, growth patterns, and cellular processes.
Interactions: The journal focuses on the interactions microorganisms have with each other, as well as their interactions with hosts or the environment. This encompasses investigations into microbial communities, symbiotic relationships, and microbial responses to different environments.
Societal significance: Nature Microbiology recognizes the societal impact of microorganisms and welcomes studies that explore their practical applications. This may include research on microbial diseases, biotechnology, or environmental remediation.
In summary, Nature Microbiology is interested in research related to the evolution, physiology and cell biology of microorganisms, their interactions, and their societal relevance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


