An atlas of genetic effects on cellular composition of the tumor microenvironment
IF 27.7
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
Abstract
Deciphering the composition of the tumor microenvironment (TME) is critical for understanding tumorigenesis and to design immunotherapies. In the present study, we mapped genetic effects on cell-type proportions using single-cell and bulk RNA sequencing data, identifying 3,494 immunity quantitative trait loci (immunQTLs) across 23 cancer types from The Cancer Genome Atlas. Functional annotation revealed regulatory potential and we further assigned 1,668 genes that regulate TME composition. We constructed a combined immunQTL map by integrating data from European and Chinese colorectal cancer (CRC) samples. A polygenic risk score that incorporates these immunQTLs and hits on a genome-wide association study outperformed in CRC risk stratification within 447,495 multiethnic individuals. Using large-scale population cohorts, we identified that the immunQTL rs1360948 is associated with CRC risk and prognosis. Mechanistically, the rs1360948-G-allele increases CCL2 expression, recruiting regulatory T cells that can exert immunosuppressive effects on CRC progression. Blocking the CCL2–CCR2 axis enhanced anti-programmed cell death protein 1 ligand therapy. Finally, we have established a database (CancerlmmunityQTL2) to serve the research community and advance our understanding of immunogenomic interactions in cancer pathogenesis. Here the authors pull apart the cellular composition of the tumor microenvironment using RNA sequencing data from a wide variety of cancer types. They identify immunity quantitative trait loci and integrate these data with analysis of colorectal cancer cohorts, creating a polygenic risk score and a database for community access.
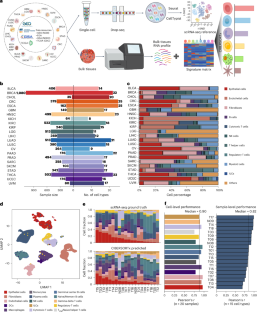
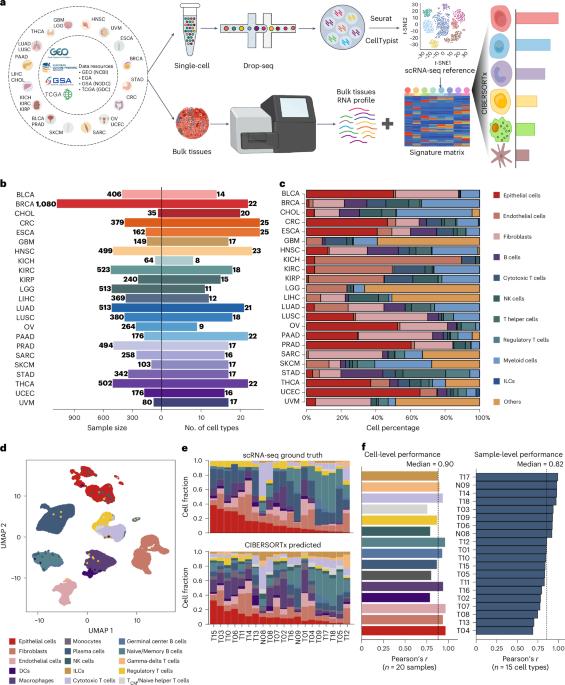
基因对肿瘤微环境细胞组成的影响图谱
解读肿瘤微环境(TME)的组成对于了解肿瘤发生和设计免疫疗法至关重要。在本研究中,我们利用单细胞和大容量 RNA 测序数据绘制了细胞类型比例的遗传效应图,在《癌症基因组图谱》的 23 种癌症类型中确定了 3494 个免疫定量性状位点(immunQTLs)。功能注释揭示了调控潜力,我们进一步分配了 1668 个调控 TME 组成的基因。我们整合了欧洲和中国结直肠癌(CRC)样本的数据,构建了综合免疫 QTL 图谱。在对 447495 名多种族个体进行 CRC 风险分层时,一个包含了这些免疫 QTL 和全基因组关联研究命中基因的多基因风险评分表现优异。通过大规模人群队列研究,我们发现免疫 QTL rs1360948 与 CRC 风险和预后有关。从机理上讲,rs1360948-G-等位基因会增加 CCL2 的表达,招募调节性 T 细胞,从而对 CRC 的进展产生免疫抑制作用。阻断 CCL2-CCR2 轴可增强抗程序性细胞死亡蛋白 1 配体疗法。最后,我们建立了一个数据库(CancerlmmunityQTL2),以服务于研究界,促进我们对癌症发病机制中免疫基因组相互作用的理解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Immunology
医学-免疫学
CiteScore
40.00
自引率
2.30%
发文量
248
审稿时长
4-8 weeks
期刊介绍:
Nature Immunology is a monthly journal that publishes the highest quality research in all areas of immunology. The editorial decisions are made by a team of full-time professional editors. The journal prioritizes work that provides translational and/or fundamental insight into the workings of the immune system. It covers a wide range of topics including innate immunity and inflammation, development, immune receptors, signaling and apoptosis, antigen presentation, gene regulation and recombination, cellular and systemic immunity, vaccines, immune tolerance, autoimmunity, tumor immunology, and microbial immunopathology. In addition to publishing significant original research, Nature Immunology also includes comments, News and Views, research highlights, matters arising from readers, and reviews of the literature. The journal serves as a major conduit of top-quality information for the immunology community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


