Structural insight into synergistic activation of human 3-methylcrotonyl-CoA carboxylase
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
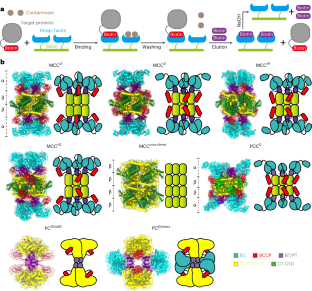
The enzymes 3-methylcrotonyl-coenzyme A (CoA) carboxylase (MCC), pyruvate carboxylase and propionyl-CoA carboxylase belong to the biotin-dependent carboxylase family located in mitochondria. They participate in various metabolic pathways in human such as amino acid metabolism and tricarboxylic acid cycle. Many human diseases are caused by mutations in those enzymes but their structures have not been fully resolved so far. Here we report an optimized purification strategy to obtain high-resolution structures of intact human endogenous MCC, propionyl-CoA carboxylase and pyruvate carboxylase in different conformational states. We also determine the structures of MCC bound to different substrates. Analysis of MCC structures in different states reveals the mechanism of the substrate-induced, multi-element synergistic activation of MCC. These results provide important insights into the catalytic mechanism of the biotin-dependent carboxylase family and are of great value for the development of new drugs for the treatment of related diseases. This work reveals structures of biotin-dependent carboxylases in different states, provides notable insight into their catalytic mechanism and may help the development of new drugs for the treatment of related diseases.

从结构上洞察人类 3-甲基巴豆酰-CoA羧化酶的协同激活作用
3-甲基巴豆酰辅酶 A(CoA)羧化酶(MCC)、丙酮酸羧化酶和丙酰-CoA 羧化酶属于生物素依赖性羧化酶家族,位于线粒体中。它们参与人类的各种代谢途径,如氨基酸代谢和三羧酸循环。许多人类疾病都是由这些酶的突变引起的,但它们的结构至今尚未完全解析。在此,我们报告了一种优化的纯化策略,以获得完整的人类内源性 MCC、丙酰-CoA 羧化酶和丙酮酸羧化酶在不同构象状态下的高分辨率结构。我们还确定了与不同底物结合的 MCC 的结构。对不同状态下 MCC 结构的分析揭示了底物诱导、多元素协同激活 MCC 的机制。这些结果为了解生物素依赖性羧化酶家族的催化机理提供了重要依据,对开发治疗相关疾病的新药具有重要价值。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


