The structural basis of pyridoxal-5′-phosphate-dependent β-NAD-alkylating enzymes
IF 42.8
1区 化学
Q1 CHEMISTRY, PHYSICAL
引用次数: 0
Abstract
SbzP is a unique pyridoxal-5′-phosphate-dependent enzyme, which catalyses a [3+2] annulation between the pyridinium ring of β-nicotinamide adenine dinucleotide (β-NAD) and an electron rich β,γ-unsaturated quinonoid derived from S-adenosylmethionine in natural product azaindane antibiotics biosynthesis. The SbzP-mediated annulation has been proposed to be a rare tandem C–C bond formation, but its structural basis and catalytic mechanism remain largely unknown. Here we report the β-NAD-complexed structure of PseP (SbzP homologue), identified by cryo-electron microscopy. Structure-based mutagenesis, stopped-flow analysis, thermal shift and surface plasmon resonance analysis identified the important residues for the substrate binding. Molecular dynamics simulations provided insights regarding how the enzyme orients the Cγ of the unsaturated quinonoid to β-NAD. In addition, density functional theory calculations confirmed that the proposed stepwise mechanism is more likely than a pericyclization mechanism. This study provides the structural basis of a pyridoxal-5′-phosphate-dependent enzyme that catalyses nucleophilic Cγ addition and β-NAD processing in natural product biosynthesis. Recently, the pyridoxal-5′-phosphate-dependent enzyme SbzP was reported to catalyse a [3+2]-annulation reaction yielding β-NAD-derived antibiotics. Now, cryo-electron microscopy structures of a stable homologue and computational simulations provide structural and mechanistic insights into this enzymatic reaction.
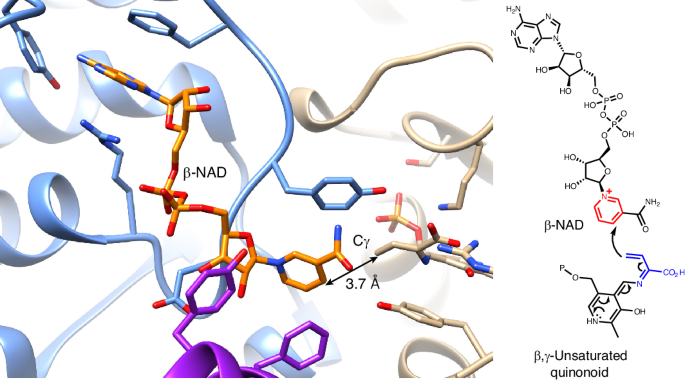
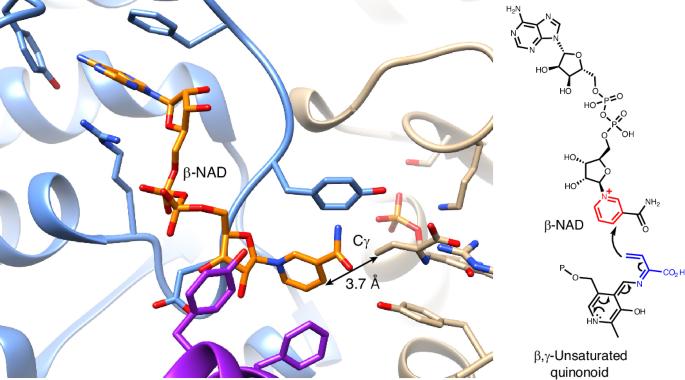
依赖吡哆醛-5′-磷酸的β-NAD-烷基化酶的结构基础
SbzP 是一种独特的依赖于吡哆醛-5′-磷酸的酶,在天然产品氮杂环丁烷抗生素的生物合成过程中,它能催化β-烟酰胺腺嘌呤二核苷酸(β-NAD)的吡啶环与来自 S-腺苷蛋氨酸的富电子β,γ-不饱和类醌之间的[3+2]环化反应。SbzP 介导的环化被认为是一种罕见的串联 C-C 键形成,但其结构基础和催化机理在很大程度上仍然未知。在此,我们报告了通过冷冻电镜鉴定的 PseP(SbzP 同源物)β-NAD 复合物结构。基于结构的诱变、停流分析、热位移和表面等离子体共振分析确定了底物结合的重要残基。分子动力学模拟揭示了酶如何将不饱和类醌的 Cγ 定向到 β-NAD。此外,密度泛函理论计算证实,所提出的逐步机制比周环化机制更有可能。这项研究为一种依赖于吡哆醛-5′-磷酸的酶提供了结构基础,这种酶在天然产物的生物合成过程中催化亲核 Cγ 加成和 β-NAD 处理。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Catalysis
Chemical Engineering-Bioengineering
CiteScore
52.10
自引率
1.10%
发文量
140
期刊介绍:
Nature Catalysis serves as a platform for researchers across chemistry and related fields, focusing on homogeneous catalysis, heterogeneous catalysis, and biocatalysts, encompassing both fundamental and applied studies. With a particular emphasis on advancing sustainable industries and processes, the journal provides comprehensive coverage of catalysis research, appealing to scientists, engineers, and researchers in academia and industry.
Maintaining the high standards of the Nature brand, Nature Catalysis boasts a dedicated team of professional editors, rigorous peer-review processes, and swift publication times, ensuring editorial independence and quality. The journal publishes work spanning heterogeneous catalysis, homogeneous catalysis, and biocatalysis, covering areas such as catalytic synthesis, mechanisms, characterization, computational studies, nanoparticle catalysis, electrocatalysis, photocatalysis, environmental catalysis, asymmetric catalysis, and various forms of organocatalysis.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


