A phosphorylation-controlled switch confers cell cycle-dependent protein relocalization
IF 17.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
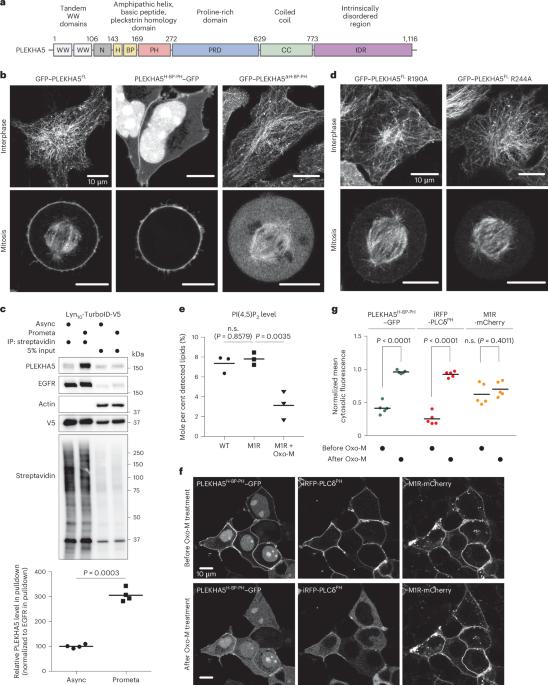
Tools for acute manipulation of protein localization enable elucidation of spatiotemporally defined functions, but their reliance on exogenous triggers can interfere with cell physiology. This limitation is particularly apparent for studying mitosis, whose highly choreographed events are sensitive to perturbations. Here we exploit the serendipitous discovery of a phosphorylation-controlled, cell cycle-dependent localization change of the adaptor protein PLEKHA5 to develop a system for mitosis-specific protein recruitment to the plasma membrane that requires no exogenous stimulus. Mitosis-enabled anchor-away/recruiter system comprises an engineered, 15 kDa module derived from PLEKHA5 capable of recruiting functional protein cargoes to the plasma membrane during mitosis, either through direct fusion or via GFP–GFP nanobody interaction. Applications of the mitosis-enabled anchor-away/recruiter system include both knock sideways to rapidly extract proteins from their native localizations during mitosis and conditional recruitment of lipid-metabolizing enzymes for mitosis-selective editing of plasma membrane lipid content, without the need for exogenous triggers or perturbative synchronization methods. Cao et al. describe the development and application of an engineered protein system (MARS) derived from PLEKHA5 that allows mitosis-specific recruitment of proteins to the plasma membrane to study protein function in cell division.
磷酸化控制的开关可实现依赖细胞周期的蛋白质重新定位
对蛋白质定位进行急性操作的工具能够阐明时空定义的功能,但它们对外源触发器的依赖会干扰细胞生理。这种局限性在研究有丝分裂时尤为明显,因为有丝分裂过程中高度编排的事件对扰动非常敏感。在这里,我们利用偶然发现的由磷酸化控制的、依赖于细胞周期的适配蛋白 PLEKHA5 的定位变化,开发了一种无需外源刺激就能将有丝分裂特异性蛋白招募到质膜上的系统。支持有丝分裂的锚移/招募系统包括一个源自 PLEKHA5 的 15 kDa 模块,该模块能够在有丝分裂期间通过直接融合或 GFP-GFP 纳米抗体相互作用将功能性蛋白质货物招募到质膜上。支持有丝分裂的锚移/招募系统的应用包括:在有丝分裂过程中侧敲,快速将蛋白质从其原生定位中提取出来;有条件地招募脂质代谢酶,对质膜脂质含量进行有丝分裂选择性编辑,而不需要外源触发器或扰动性同步方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


