Rare coding variant analysis for human diseases across biobanks and ancestries
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
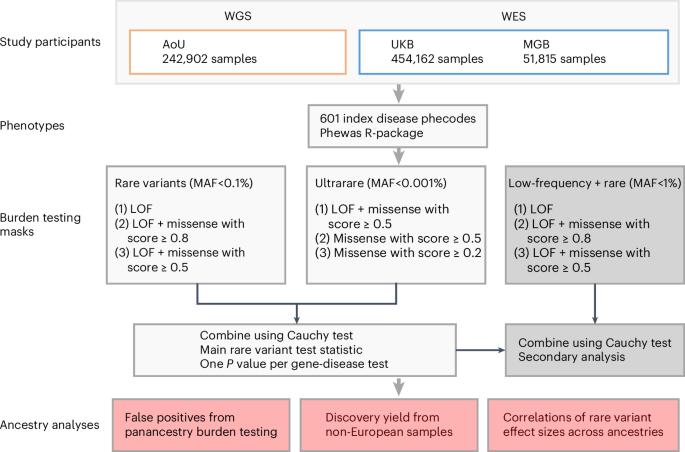
Large-scale sequencing has enabled unparalleled opportunities to investigate the role of rare coding variation in human phenotypic variability. Here, we present a pan-ancestry analysis of sequencing data from three large biobanks, including the All of Us research program. Using mixed-effects models, we performed gene-based rare variant testing for 601 diseases across 748,879 individuals, including 155,236 with ancestry dissimilar to European. We identified 363 significant associations, which highlighted core genes for the human disease phenome and identified potential novel associations, including UBR3 for cardiometabolic disease and YLPM1 for psychiatric disease. Pan-ancestry burden testing represented an inclusive and useful approach for discovery in diverse datasets, although we also highlight the importance of ancestry-specific sensitivity analyses in this setting. Finally, we found that effect sizes for rare protein-disrupting variants were concordant between samples similar to European ancestry and other genetic ancestries (βDeming = 0.7–1.0). Our results have implications for multi-ancestry and cross-biobank approaches in sequencing association studies for human disease. Gene-based rare variant analyses for 601 diseases across 748,879 individuals from three biobanks identify 363 significant associations and highlight important considerations for multi-ancestry and cross-biobank sequencing studies.
跨生物库和祖先的人类疾病罕见编码变异分析
大规模测序为研究罕见编码变异在人类表型变异中的作用提供了无与伦比的机会。在此,我们对来自三个大型生物库(包括 "我们所有人 "研究计划)的测序数据进行了泛种群分析。利用混合效应模型,我们对 748,879 人的 601 种疾病进行了基于基因的罕见变异测试,其中包括 155,236 名祖先与欧洲人不同的个体。我们发现了 363 个重大关联,突出了人类疾病表型的核心基因,并发现了潜在的新型关联,包括与心脏代谢疾病相关的 UBR3 和与精神疾病相关的 YLPM1。尽管我们也强调了在这种情况下进行特定祖先敏感性分析的重要性,但泛祖先负担测试是在不同数据集中进行发现的一种包容而有用的方法。最后,我们发现,在类似欧洲血统的样本和其他遗传血统的样本之间,罕见蛋白质干扰变体的效应大小是一致的(βDeming = 0.7-1.0)。我们的研究结果对人类疾病测序关联研究中的多血统和跨生物库方法具有重要意义。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


