A deep-learning framework for enhancing habitat identification based on species composition
Abstract
Aims
The accurate classification of habitats is essential for effective biodiversity conservation. The goal of this study was to harness the potential of deep learning to advance habitat identification in Europe. We aimed to develop and evaluate models capable of assigning vegetation-plot records to the habitats of the European Nature Information System (EUNIS), a widely used reference framework for European habitat types.
Location
The framework was designed for use in Europe and adjacent areas (e.g., Anatolia, Caucasus).
Methods
We leveraged deep-learning techniques, such as transformers (i.e., models with attention components able to learn contextual relations between categorical and numerical features) that we trained using spatial k-fold cross-validation (CV) on vegetation plots sourced from the European Vegetation Archive (EVA), to show that they have great potential for classifying vegetation-plot records. We tested different network architectures, feature encodings, hyperparameter tuning and noise addition strategies to identify the optimal model. We used an independent test set from the National Plant Monitoring Scheme (NPMS) to evaluate its performance and compare its results against the traditional expert systems.
Results
Exploration of the use of deep learning applied to species composition and plot-location criteria for habitat classification led to the development of a framework containing a wide range of models. Our selected algorithm, applied to European habitat types, significantly improved habitat classification accuracy, achieving a more than twofold improvement compared to the previous state-of-the-art (SOTA) method on an external data set, clearly outperforming expert systems. The framework is shared and maintained through a GitHub repository.
Conclusions
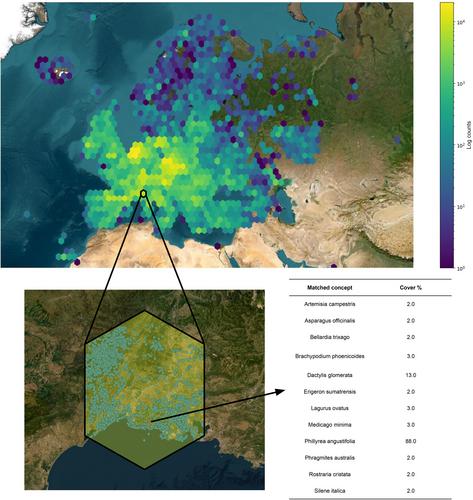
Our results demonstrate the potential benefits of the adoption of deep learning for improving the accuracy of vegetation classification. They highlight the importance of incorporating advanced technologies into habitat monitoring. These algorithms have shown to be better suited for habitat type prediction than expert systems. They push the accuracy score on a database containing hundreds of thousands of standardized presence/absence European surveys to 88.74%, as assessed by expert judgment. Finally, our results showcase that species dominance is a strong marker of ecosystems and that the exact cover abundance of the flora is not required to train neural networks with predictive performances. The framework we developed can be used by researchers and practitioners to accurately classify habitats.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


