A unified framework to investigate and interpret hybrid and allopolyploid biodiversity across biological scales
Abstract
Premise
Hybridization and polyploidization are common in vascular plants and important drivers of biodiversity by facilitating speciation and ecological diversification. A primary limitation to making broad synthetic discoveries in hybrid and allopolyploid biodiversity research is the absence of a standardized framework to compare data across studies and biological scales.
Methods
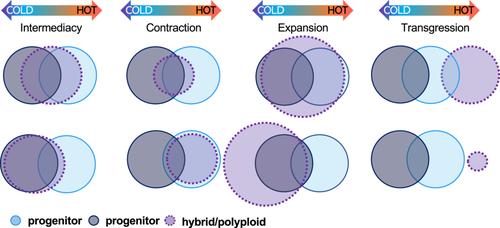
Here, I present a new quantitative framework to investigate and interpret patterns in hybrid and allopolyploid biology called the divergence index (DI). The DI framework produces standardized data that are comparable across studies and variables. To show how the DI framework can be used to synthesize data, I analyzed published biochemical, physiological, and ecological trait data of hybrids and allopolyploids. I also apply key ecological and evolutionary concepts in hybrid and polyploid biology to translate nominal outcomes, including transgression, intermediacy, expansion, and contraction, in continuous DI space.
Results
Biochemical, physiological, ecological, and evolutionary data can all be analyzed, visualized, and interpreted in the DI framework. The DI framework is particularly suited to standardize and compare variables with very different scales. When using the DI framework to understand niche divergence, a metric of niche overlap can be used to complement insights to centroid and breadth changes.
Discussion
The DI framework is an accessible framework for hybrid and allopolyploid biology and represents a flexible and intuitive tool that can be used to reconcile outstanding problems in plant biodiversity research.


 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


