Deciphering the role of oxidative stress genes in idiopathic pulmonary fibrosis: a multi-omics mendelian randomization approach
IF 4.5
3区 医学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
Oxidative stress (OS) is crucial in idiopathic pulmonary fibrosis (IPF) pathogenesis, with its genes potentially acting as both causes and consequences of the disease. We identified OS-related genes from GeneCards and performed a meta-analysis on pulmonary transcriptome datasets to discover differentially expressed genes (DEGs) related to OS in IPF. We integrated this data with the largest available IPF GWAS summaries, expression quantitative trait loci (eQTLs), and DNA methylation QTLs (mQTLs) from blood. This approach aimed to identify blood OS genes and regulatory elements linked to IPF risk, incorporating the latest pulmonary eQTLs and bronchoalveolar lavage fluid microbial QTLs (bmQTLs) for a comprehensive view of gene-lung microbiota interactions through SMR and colocalization analyses. Sensitivity analyses were conducted using two additional mendelian randomization (MR) methods. Meta-analysis revealed 1090 differentially expressed OS genes between IPF patients and controls. Integration with IPF GWAS, eQTL, and mQTL data identified key genes and regulatory elements involved in IPF pathogenesis, highlighting the role of specific genes such as KCNMA1 and SLC22A5 in modulating IPF risk through epigenetic mechanisms. Colocalization analysis further identified potential interactions between gene expression and lung microbiota. Our findings elucidate the complex interplay between OS genes and IPF, suggesting potential therapeutic targets and highlighting the importance of considering epigenetic and microbial interactions in the disease’s etiology and progression.
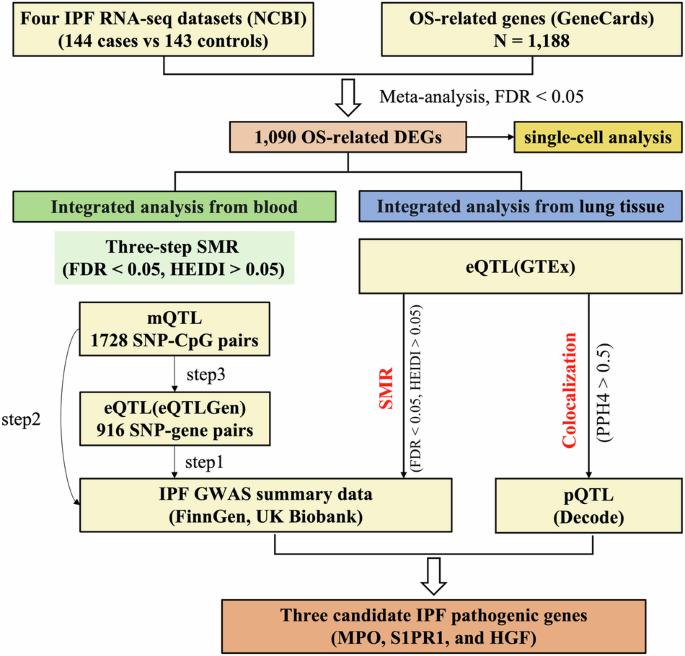
解密氧化应激基因在特发性肺纤维化中的作用:一种多组学孟德尔随机方法。
氧化应激(OS)在特发性肺纤维化(IPF)发病机制中至关重要,其基因可能既是疾病的原因,也是疾病的后果。我们从基因卡片(GeneCards)中确定了与OS相关的基因,并对肺转录组数据集进行了荟萃分析,以发现与IPF中OS相关的差异表达基因(DEGs)。我们将这些数据与现有最大的 IPF GWAS 摘要、表达定量性状位点(eQTL)和血液中的 DNA 甲基化 QTL(mQTL)进行了整合。这种方法旨在确定与 IPF 风险相关的血液 OS 基因和调控元件,并结合最新的肺 eQTL 和支气管肺泡灌洗液微生物 QTL(bmQTL),通过 SMR 和共定位分析全面了解基因与肺微生物区系的相互作用。另外还使用两种泯灭随机(MR)方法进行了敏感性分析。元分析显示,IPF 患者和对照组之间有 1090 个不同表达的 OS 基因。与 IPF GWAS、eQTL 和 mQTL 数据相结合,确定了参与 IPF 发病机制的关键基因和调控元件,突出了 KCNMA1 和 SLC22A5 等特定基因在通过表观遗传机制调节 IPF 风险方面的作用。共定位分析进一步确定了基因表达与肺部微生物群之间潜在的相互作用。我们的研究结果阐明了OS基因与IPF之间复杂的相互作用,提出了潜在的治疗靶点,并强调了考虑表观遗传学和微生物相互作用在该疾病的病因学和进展中的重要性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Genes and immunity
医学-免疫学
CiteScore
8.90
自引率
4.00%
发文量
28
审稿时长
6-12 weeks
期刊介绍:
Genes & Immunity emphasizes studies investigating how genetic, genomic and functional variations affect immune cells and the immune system, and associated processes in the regulation of health and disease. It further highlights articles on the transcriptional and posttranslational control of gene products involved in signaling pathways regulating immune cells, and protective and destructive immune responses.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


