Genome-scale quantification and prediction of pathogenic stop codon readthrough by small molecules
IF 29
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
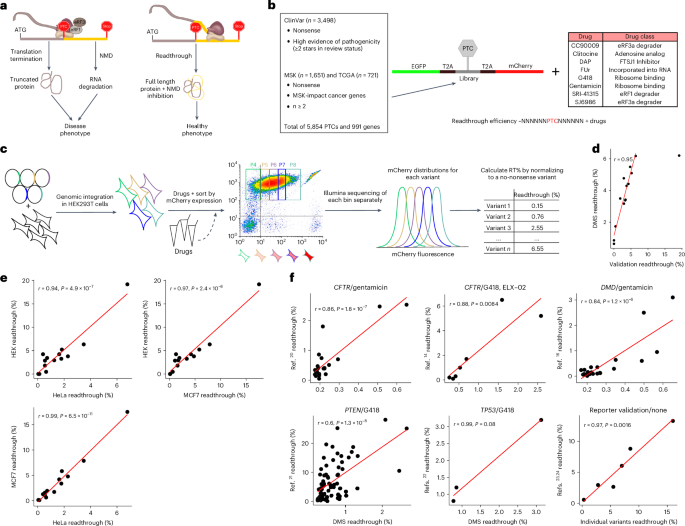
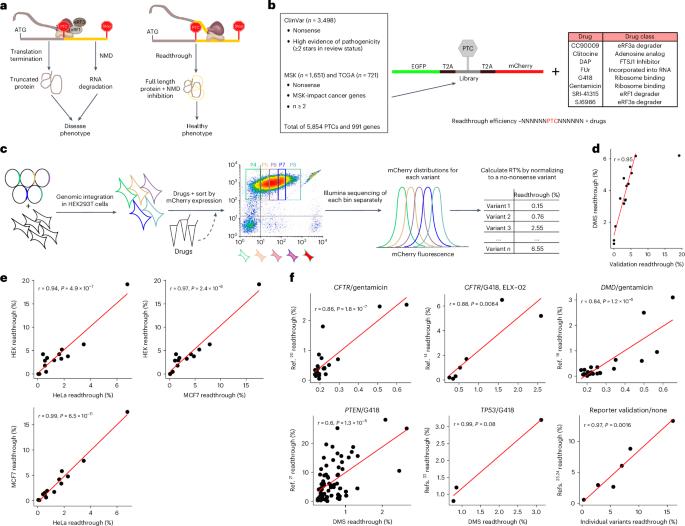
Premature termination codons (PTCs) cause ~10–20% of inherited diseases and are a major mechanism of tumor suppressor gene inactivation in cancer. A general strategy to alleviate the effects of PTCs would be to promote translational readthrough. Nonsense suppression by small molecules has proven effective in diverse disease models, but translation into the clinic is hampered by ineffective readthrough of many PTCs. Here we directly tackle the challenge of defining drug efficacy by quantifying the readthrough of ~5,800 human pathogenic stop codons by eight drugs. We find that different drugs promote the readthrough of complementary subsets of PTCs defined by local sequence context. This allows us to build interpretable models that accurately predict drug-induced readthrough genome-wide, and we validate these models by quantifying endogenous stop codon readthrough. Accurate readthrough quantification and prediction will empower clinical trial design and the development of personalized nonsense suppression therapies. Small molecules can promote translational readthrough of premature termination codons, reducing their pathological effect. This study quantifies the readthrough of ~5,800 human pathogenic stop codons by eight drugs and builds models to predict drug-induced readthrough genome-wide.
基因组尺度量化和预测小分子致病终止密码子读穿
过早终止密码子(PTC)导致约 10-20% 的遗传性疾病,也是癌症中抑癌基因失活的主要机制。减轻过早终止密码子影响的一般策略是促进翻译通读。小分子的有义抑制已被证明在多种疾病模型中有效,但由于许多 PTCs 的无效通读,将其转化为临床应用受到了阻碍。在这里,我们通过量化八种药物对约 5,800 个人类致病终止密码子的读通情况,直接解决了界定药物疗效的难题。我们发现,不同的药物会促进由局部序列上下文定义的 PTC 互补子集的读通。这使我们能够建立可解释的模型,准确预测药物诱导的全基因组读通,我们还通过量化内源性终止密码子读通验证了这些模型。准确的读穿量化和预测将有助于临床试验设计和个性化无意义抑制疗法的开发。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


