Profiling cell identity and tissue architecture with single-cell and spatial transcriptomics
IF 81.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Single-cell transcriptomics has broadened our understanding of cellular diversity and gene expression dynamics in healthy and diseased tissues. Recently, spatial transcriptomics has emerged as a tool to contextualize single cells in multicellular neighbourhoods and to identify spatially recurrent phenotypes, or ecotypes. These technologies have generated vast datasets with targeted-transcriptome and whole-transcriptome profiles of hundreds to millions of cells. Such data have provided new insights into developmental hierarchies, cellular plasticity and diverse tissue microenvironments, and spurred a burst of innovation in computational methods for single-cell analysis. In this Review, we discuss recent advancements, ongoing challenges and prospects in identifying and characterizing cell states and multicellular neighbourhoods. We discuss recent progress in sample processing, data integration, identification of subtle cell states, trajectory modelling, deconvolution and spatial analysis. Furthermore, we discuss the increasing application of deep learning, including foundation models, in analysing single-cell and spatial transcriptomics data. Finally, we discuss recent applications of these tools in the fields of stem cell biology, immunology, and tumour biology, and the future of single-cell and spatial transcriptomics in biological research and its translation to the clinic. Single-cell and spatial transcriptomics are transforming our understanding of cell plasticity and tissue diversity. This Review discusses technical and computational advancements and challenges in characterizing cell states and tissues during embryogenesis, tumorigenesis and immune responses, and the application of these tools to the clinic.
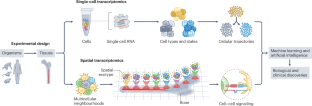

利用单细胞和空间转录组学分析细胞特征和组织结构
单细胞转录组学拓宽了我们对细胞多样性以及健康和患病组织中基因表达动态的了解。最近,空间转录组学已成为一种工具,可用于确定多细胞邻域中单细胞的背景,以及识别空间重复出现的表型或生态型。这些技术产生了大量数据集,包括数百至数百万个细胞的靶向转录组和全转录组图谱。这些数据提供了对发育层次、细胞可塑性和不同组织微环境的新见解,并推动了单细胞分析计算方法的创新。在本综述中,我们将讨论在识别和描述细胞状态和多细胞邻域方面的最新进展、持续挑战和前景。我们将讨论样本处理、数据整合、微妙细胞状态识别、轨迹建模、解卷积和空间分析等方面的最新进展。此外,我们还讨论了深度学习(包括基础模型)在单细胞和空间转录组学数据分析中越来越多的应用。最后,我们讨论了这些工具在干细胞生物学、免疫学和肿瘤生物学领域的最新应用,以及单细胞和空间转录组学在生物研究中的未来及其在临床中的应用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
CiteScore
173.60
自引率
0.50%
发文量
118
审稿时长
6-12 weeks
期刊介绍:
Nature Reviews Molecular Cell Biology is a prestigious journal that aims to be the primary source of reviews and commentaries for the scientific communities it serves. The journal strives to publish articles that are authoritative, accessible, and enriched with easily understandable figures, tables, and other display items. The goal is to provide an unparalleled service to authors, referees, and readers, and the journal works diligently to maximize the usefulness and impact of each article. Nature Reviews Molecular Cell Biology publishes a variety of article types, including Reviews, Perspectives, Comments, and Research Highlights, all of which are relevant to molecular and cell biologists. The journal's broad scope ensures that the articles it publishes reach the widest possible audience.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


