A PRE loop at the dac locus acts as a topological chromatin structure that restricts and specifies enhancer–promoter communication
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Three-dimensional (3D) genome folding has a fundamental role in the regulation of developmental genes by facilitating or constraining chromatin interactions between cis-regulatory elements (CREs). Polycomb response elements (PREs) are a specific kind of CRE involved in the memory of transcriptional states in Drosophila melanogaster. PREs act as nucleation sites for Polycomb group (PcG) proteins, which deposit the repressive histone mark H3K27me3, leading to the formation of a class of topologically associating domain (TAD) called a Polycomb domain. PREs can establish looping contacts that stabilize the gene repression of key developmental genes during development. However, the mechanism by which PRE loops fine-tune gene expression is unknown. Using clustered regularly interspaced short palindromic repeats and Cas9 genome engineering, we specifically perturbed PRE contacts or enhancer function and used complementary approaches including 4C-seq, Hi-C and Hi-M to analyze how chromatin architecture perturbation affects gene expression. Our results suggest that the PRE loop at the dac gene locus acts as a constitutive 3D chromatin scaffold during Drosophila development that forms independently of gene expression states and has a versatile function; it restricts enhancer–promoter communication and contributes to enhancer specificity. Combining genome engineering, epigenomics and multiplex three-dimensional microscopy approaches, the authors show that PRE chromatin loops form a topological scaffold, restricting promoter–enhancer communication and contributing to enhancer–promoter specificity.
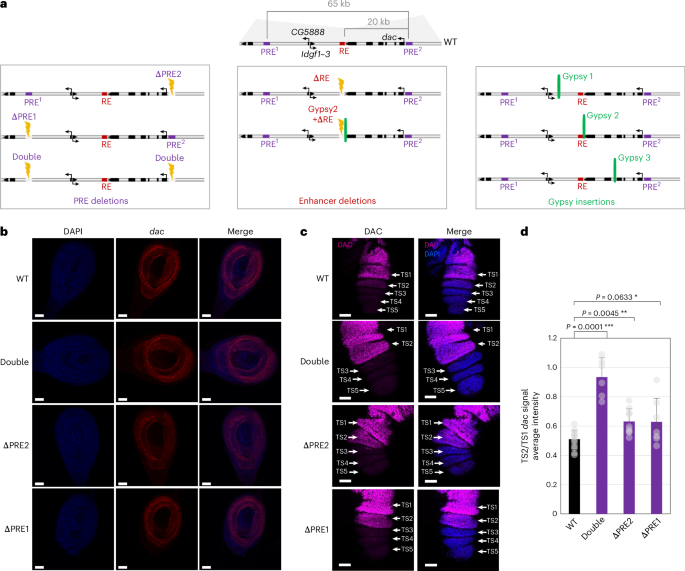

dac 基因座上的 PRE 环是一种拓扑染色质结构,它限制并指定了增强子与启动子之间的交流
三维(3D)基因组折叠通过促进或限制顺式调控元件(CRE)之间的染色质相互作用,在调控发育基因方面发挥着重要作用。多聚酶反应元件(PRE)是一种特殊的 CRE,参与了黑腹果蝇转录状态的记忆。PREs是多角体群(PcG)蛋白的成核位点,PcG蛋白会沉积抑制性组蛋白标记H3K27me3,从而形成一类拓扑关联结构域(TAD),即多角体结构域。PRE可建立环状联系,在发育过程中稳定关键发育基因的基因抑制。然而,PRE环路对基因表达进行微调的机制尚不清楚。我们利用聚类规则间隔短回文重复序列(clustered regularly interspaced short palindromic repeats)和Cas9基因组工程,特异性地扰乱了PRE接触或增强子功能,并使用4C-seq、Hi-C和Hi-M等互补方法分析了染色质结构扰动如何影响基因表达。我们的研究结果表明,在果蝇的发育过程中,dac 基因座上的 PRE 环是一个组成型三维染色质支架,它的形成与基因表达状态无关,具有多功能性;它限制增强子与启动子之间的通讯,并有助于增强子的特异性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


