Insilico sequence-structure based analysis of bacterial chromate reductase to unravel enzymatic specificity towards chromium pollution
Abstract
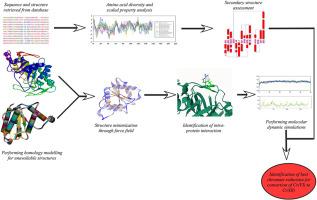
Extensive use of chromium in industrial sectors leads to hazardous consequences of chromium pollution. The hexavalent chromium is much more damaging to biological systems than the trivalent and other forms. The best strategy for bioconversion of such toxic hexavalent chromium to less toxic trivalent chromium is the enzymatic breakdown using chromate reductase produced by several bacteria. The employment of in silico analysis aids in the quick detection of a protein's function and stability. Therefore, an insilico sequence and structure analysis of chromate reductase of the different bacterial groups has been performed. Amino acid variation reveals more charged polar residues in the extremophilic group than the mesophiles and is more hydrophilic. According to secondary structure data, beta-bulge formation was only observed in Thermus scotoductus. A higher abundance of network salt bridges and isolated aromatic-sulphur interactions were observed in extremophiles, indicating higher stability. Rhombus network salt bridge formation is a novel finding in thermophilic bacteria. Better isolated cation-pi interactions were observed in both thermophiles and halophiles. Molecular dynamics simulation studies through RMSD observed similar trajectories between thermophilic and psychrophilic proteins indicating a shared structural pattern. RMSF studies indicate the lowest fluctuations by thermophiles compared to other groups. Rg values indicate that chromate reductase of Halomonas subglaciescola had the lowest plot indicating its most compactness. The chromate reductase from psychrophilic Psychrobacillus psychrotolerans displayed the lowest SASA, indicating the most stability. Hydrogen bond number was also higher in extremophiles. This is the first report on such comparative analysis of chromate reductase from extremophiles indicating that they are more preferred candidates in providing a more stable enzyme for better chromium remediation.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


