Deep Learning‐Based Classification of Histone–DNA Interactions Using Drying Droplet Patterns
IF 3.5
2区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Developing scalable and accurate predictive analytical methods for the classification of protein‐DNA binding is critical for advancing our understanding of molecular biology, disease mechanisms, and a wide spectrum of biotechnological and medical applications. It is discovered that histone–DNA interactions can be stratified based on stain patterns created by the deposition of various nucleoprotein solutions onto a substrate. In this study, a deep‐learning neural network is applied to categorize polarized light microscopy images of drying droplet deposits originating from different histone–DNA mixtures. These DNA stain patterns featured high reproducibility across different species and thus enabled comprehensive DNA categorization (100% accuracy) and accurate prediction of their respective binding affinities to histones. Eukaryotic DNA, which has a higher binding affinity to mammalian histones than prokaryotic DNA, is associated with a higher overall prediction accuracy. For a given species, the average prediction accuracy increased with DNA size. To demonstrate generalizability, a pre‐trained CNN is challenged with unknown images that originated from DNA samples of species not included in the training set. The CNN classified these unknown histone‐DNA samples as either strong or medium binders with 84.4% and 96.25% accuracy, respectively.
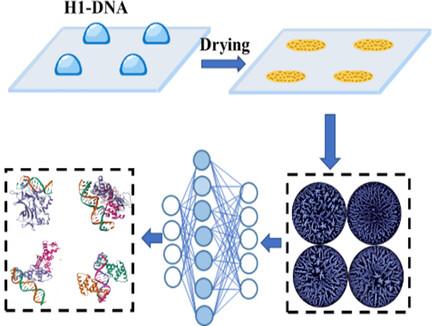
利用干燥液滴模式对组蛋白-DNA相互作用进行基于深度学习的分类
为蛋白质-DNA 结合的分类开发可扩展且准确的预测分析方法,对于促进我们对分子生物学、疾病机理以及广泛的生物技术和医学应用的理解至关重要。研究发现,组蛋白与 DNA 的相互作用可根据各种核蛋白溶液沉积在基底上形成的染色模式进行分层。在这项研究中,深度学习神经网络被用于对源自不同组蛋白-DNA 混合物的干燥液滴沉积的偏振光显微镜图像进行分类。这些DNA染色模式在不同物种之间具有很高的可重复性,因此能够进行全面的DNA分类(准确率为100%),并准确预测它们各自与组蛋白的结合亲和力。与原核 DNA 相比,真核 DNA 与哺乳动物组蛋白的结合亲和力更高,因此总体预测准确率也更高。对于特定物种,平均预测准确率随 DNA 大小的增加而提高。为了证明其通用性,预先训练好的 CNN 要面对来自未列入训练集的物种 DNA 样本的未知图像的挑战。CNN 将这些未知的组蛋白 DNA 样本分类为强粘合剂或中等粘合剂,准确率分别为 84.4% 和 96.25%。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

ACS Chemical Biology
生物-生化与分子生物学
CiteScore
7.50
自引率
5.00%
发文量
353
审稿时长
3.3 months
期刊介绍:
ACS Chemical Biology provides an international forum for the rapid communication of research that broadly embraces the interface between chemistry and biology.
The journal also serves as a forum to facilitate the communication between biologists and chemists that will translate into new research opportunities and discoveries. Results will be published in which molecular reasoning has been used to probe questions through in vitro investigations, cell biological methods, or organismic studies.
We welcome mechanistic studies on proteins, nucleic acids, sugars, lipids, and nonbiological polymers. The journal serves a large scientific community, exploring cellular function from both chemical and biological perspectives. It is understood that submitted work is based upon original results and has not been published previously.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


