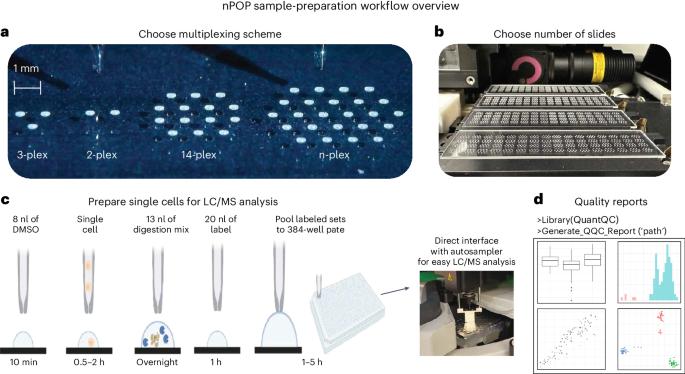
Massively parallel sample preparation for multiplexed single-cell proteomics using nPOP
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Single-cell proteomics by mass spectrometry (MS) allows the quantification of proteins with high specificity and sensitivity. To increase its throughput, we developed nano-proteomic sample preparation (nPOP), a method for parallel preparation of thousands of single cells in nanoliter-volume droplets deposited on glass slides. Here, we describe its protocol with emphasis on its flexibility to prepare samples for different multiplexed MS methods. An implementation using the plexDIA MS multiplexing method, which uses non-isobaric mass tags to barcode peptides from different samples for data-independent acquisition, demonstrates accurate quantification of ~3,000–3,700 proteins per human cell. A separate implementation with isobaric mass tags and prioritized data acquisition demonstrates analysis of 1,827 single cells at a rate of >1,000 single cells per day at a depth of 800–1,200 proteins per human cell. The protocol is implemented by using a cell-dispensing and liquid-handling robot—the CellenONE instrument—and uses readily available consumables, which should facilitate broad adoption. nPOP can be applied to all samples that can be processed to a single-cell suspension. It takes 1 or 2 d to prepare >3,000 single cells. We provide metrics and software (the QuantQC R package) for quality control and data exploration. QuantQC supports the robust scaling of nPOP to higher plex reagents for achieving reliable and scalable single-cell proteomics. nPOP is a method for parallel preparation of thousands of single cells in nanoliter-volume droplets deposited on glass slides by using the CellenONE instrument. This protocol describes the liquid handling for multiplexed mass spectrometry proteomics.
利用 nPOP 进行大规模并行样品制备,实现多重单细胞蛋白质组学。
通过质谱(MS)进行单细胞蛋白质组学研究,可以对蛋白质进行高特异性和高灵敏度的定量分析。为了提高通量,我们开发了纳米蛋白质组样品制备(nPOP),这是一种在玻璃载玻片上沉积的纳升体积液滴中平行制备数千个单细胞的方法。在此,我们介绍了该方法的操作规程,重点是它在制备用于不同多重质谱方法的样品时的灵活性。使用 plexDIA MS 多路复用方法(该方法使用非等位质量标签对来自不同样品的肽进行条形码编码,以进行独立于数据的采集)的实施表明,每个人体细胞可精确定量约 3,000-3,700 个蛋白质。另一项使用等压质量标签和优先数据采集的实施方案显示,以每天大于 1,000 个单细胞的速度分析了 1,827 个单细胞,每个人体细胞的蛋白质深度为 800-1,200 个。nPOP 可用于所有可处理成单细胞悬液的样本。制备大于 3,000 个单细胞只需 1 或 2 天。我们提供用于质量控制和数据探索的指标和软件(QuantQC R 软件包)。QuantQC 支持 nPOP 向更高复合试剂的稳健扩展,以实现可靠、可扩展的单细胞蛋白质组学。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


