Structural basis of RfaH-mediated transcription–translation coupling
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
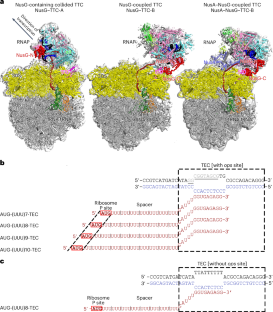
The NusG paralog RfaH mediates bacterial transcription–translation coupling in genes that contain a DNA sequence element, termed an ops site, required for pausing RNA polymerase (RNAP) and for loading RfaH onto the paused RNAP. Here, we report cryo-electron microscopy structures of transcription–translation complexes (TTCs) containing Escherichia coli RfaH. The results show that RfaH bridges RNAP and the ribosome, with the RfaH N-terminal domain interacting with RNAP and the RfaH C-terminal domain interacting with the ribosome. The results show that the distribution of translational and orientational positions of RNAP relative to the ribosome in RfaH-coupled TTCs is more restricted than in NusG-coupled TTCs because of the more restricted flexibility of the RfaH interdomain linker. The results further suggest that the structural organization of RfaH-coupled TTCs in the ‘loading state’, in which RNAP and RfaH are located at the ops site during formation of the TTC, is the same as the structural organization of RfaH-coupled TTCs in the ‘loaded state’, in which RNAP and RfaH are located at positions downstream of the ops site during function of the TTC. The results define the structural organization of RfaH-containing TTCs and set the stage for analysis of functions of RfaH during translation initiation and transcription–translation coupling. Here, the authors report cryo-electron microscopy structures of Escherichia coli transcription–translation complexes containing the transcription–translation coupling factor RfaH, showing that RfaH physically bridges RNA polymerase and the ribosome.

RfaH 介导的转录-翻译耦合的结构基础
NusG Paralog RfaH 在含有 DNA 序列元件(称为 ops 位点)的基因中介导细菌转录-翻译耦合,该 DNA 序列元件是暂停 RNA 聚合酶(RNAP)和将 RfaH 装载到暂停的 RNAP 上所必需的。在此,我们报告了含有大肠杆菌 RfaH 的转录-翻译复合物(TTC)的冷冻电镜结构。结果表明,RfaH是RNAP和核糖体的桥梁,RfaH的N端结构域与RNAP相互作用,RfaH的C端结构域与核糖体相互作用。结果表明,在 RfaH 耦合的 TTC 中,RNAP 相对于核糖体的平移和定向位置分布比在 NusG 耦合的 TTC 中更受限制,因为 RfaH 域间连接器的灵活性更受限制。研究结果进一步表明,RfaH-偶联 TTC 在 "加载状态 "下的结构组织与 RfaH-偶联 TTC 在 "加载状态 "下的结构组织是相同的,在 "加载状态 "下,RNAP 和 RfaH 位于 TTC 功能的 ops 位点下游。这些结果确定了含 RfaH 的 TTC 的结构组织,并为分析 RfaH 在翻译启动和转录-翻译耦合过程中的功能奠定了基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


