Spindle architecture constrains karyotype evolution
IF 17.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
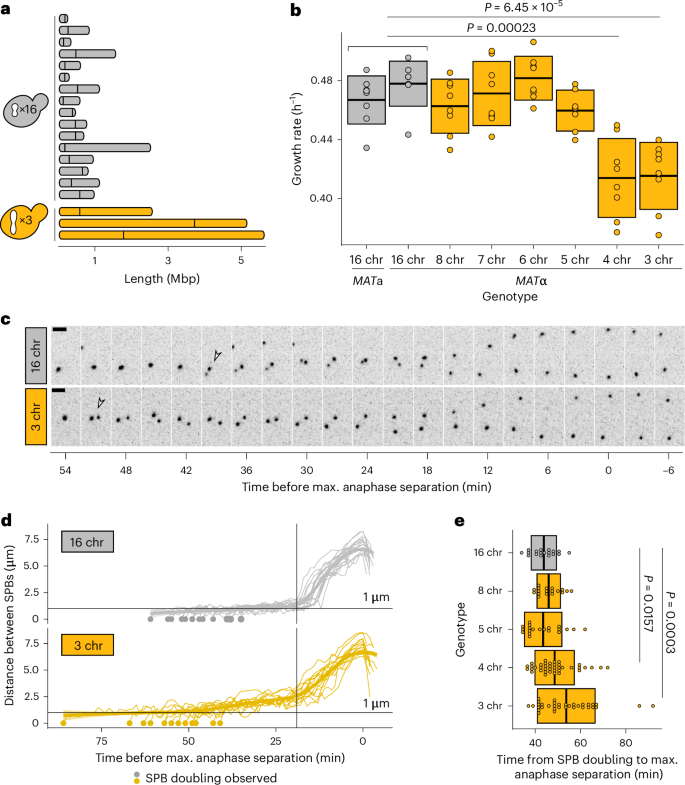
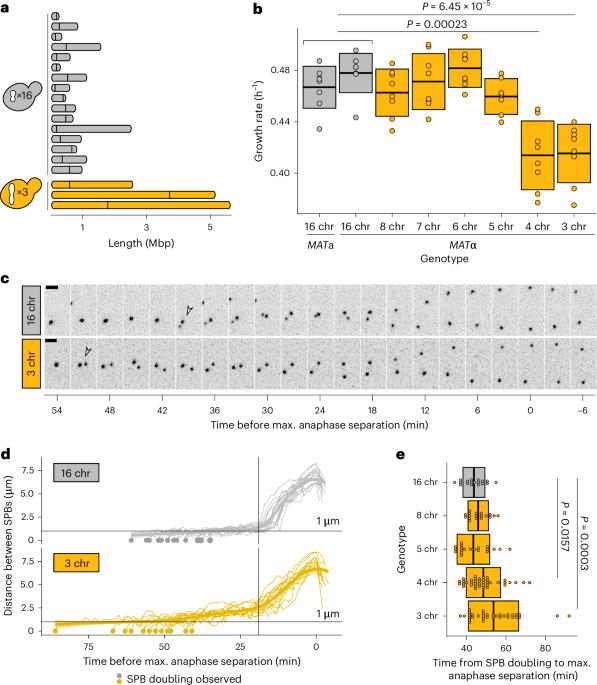
The eukaryotic cell division machinery must rapidly and reproducibly duplicate and partition the cell’s chromosomes in a carefully coordinated process. However, chromosome numbers vary dramatically between genomes, even on short evolutionary timescales. We sought to understand how the mitotic machinery senses and responds to karyotypic changes by using a series of budding yeast strains in which the native chromosomes have been successively fused. Using a combination of cell biological profiling, genetic engineering and experimental evolution, we show that chromosome fusions are well tolerated up until a critical point. Cells with fewer than five centromeres lack the necessary number of kinetochore-microtubule attachments needed to counter outward forces in the metaphase spindle, triggering the spindle assembly checkpoint and prolonging metaphase. Our findings demonstrate that spindle architecture is a constraining factor for karyotype evolution. Helsen et al. use experimental evolution and chromosome engineering to probe the link between karyotype changes and the cell division machinery. They conclude that spindle organization dictates the available trajectories for karyotype evolution.
纺锤体结构制约核型进化
真核细胞的分裂机制必须在一个精心协调的过程中快速、可重复地复制和分割细胞的染色体。然而,即使在短进化时间尺度上,不同基因组之间的染色体数目也会发生巨大变化。我们试图利用一系列原生染色体连续融合的萌发酵母菌株,了解有丝分裂机制如何感知核型变化并做出反应。我们综合运用细胞生物学分析、基因工程和实验进化等方法,证明染色体融合在临界点之前都能很好地容忍。中心粒少于五个的细胞缺乏必要数量的动点核-微管连接,无法抵消分裂期纺锤体的外力,从而触发纺锤体组装检查点并延长分裂期。我们的研究结果表明,纺锤体结构是核型进化的一个制约因素。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


