Structural basis for processive daughter-strand synthesis and proofreading by the human leading-strand DNA polymerase Pol ε
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
During chromosome replication, the nascent leading strand is synthesized by DNA polymerase epsilon (Pol ε), which associates with the sliding clamp processivity factor proliferating cell nuclear antigen (PCNA) to form a processive holoenzyme. For high-fidelity DNA synthesis, Pol ε relies on nucleotide selectivity and its proofreading ability to detect and excise a misincorporated nucleotide. Here, we present cryo-electron microscopy (cryo-EM) structures of human Pol ε in complex with PCNA, DNA and an incoming nucleotide, revealing how Pol ε associates with PCNA through its PCNA-interacting peptide box and additional unique features of its catalytic domain. Furthermore, by solving a series of cryo-EM structures of Pol ε at a mismatch-containing DNA, we elucidate how Pol ε senses and edits a misincorporated nucleotide. Our structures delineate steps along an intramolecular switching mechanism between polymerase and exonuclease activities, providing the basis for a proofreading mechanism in B-family replicative polymerases. Using cryo-electron microscopy, the authors deepen our mechanistic understanding of nascent leading-strand synthesis during human DNA replication and provide the basis for a proofreading mechanism in B-family replicative polymerases.
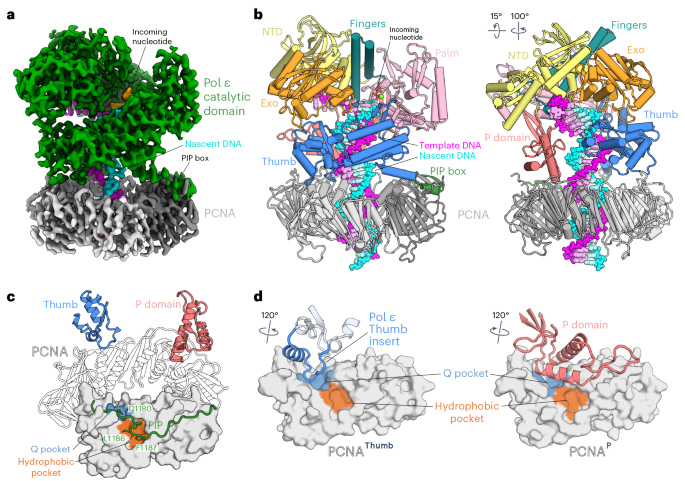

人类前导链 DNA 聚合酶 Pol ε 进行子链合成和校对的结构基础
在染色体复制过程中,新生前导链由 DNA 聚合酶ε(Pol ε)合成,它与滑动钳加工因子增殖细胞核抗原(PCNA)结合形成一个加工全酶。Pol ε依靠核苷酸选择性及其校对能力来检测和切除错误结合的核苷酸,从而实现高保真的DNA合成。在这里,我们展示了人Pol ε与PCNA、DNA和输入核苷酸复合物的冷冻电子显微镜(cryo-EM)结构,揭示了Pol ε如何通过其PCNA-interacting肽盒与PCNA结合,以及其催化结构域的其他独特特征。此外,通过解决 Pol ε 在含错配 DNA 上的一系列低温电子显微镜结构,我们阐明了 Pol ε 如何感知和编辑误入的核苷酸。我们的结构描述了聚合酶和外切酶活性之间分子内切换机制的步骤,为 B-家族复制聚合酶的校对机制提供了基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


