Reference genome sequence and population genomic analysis of peas provide insights into the genetic basis of Mendelian and other agronomic traits
IF 31.7
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
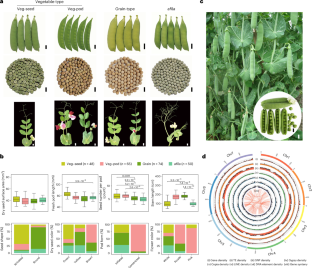
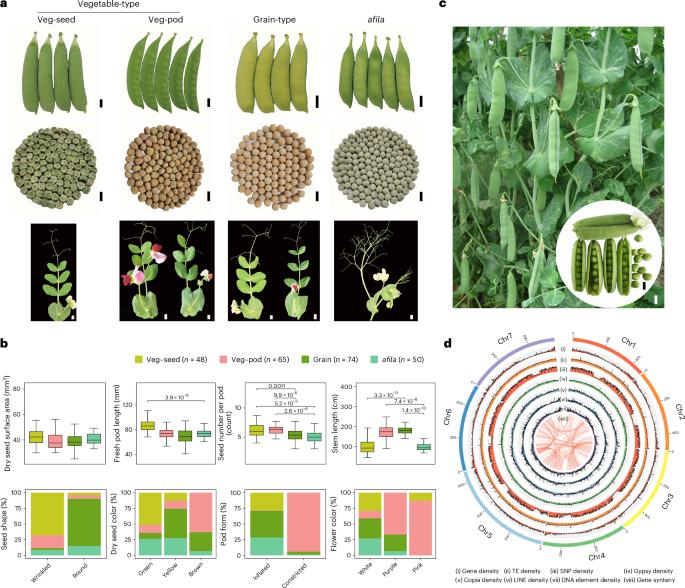
Peas are essential for human nutrition and played a crucial role in the discovery of Mendelian laws of inheritance. In this study, we assembled the genome of the elite vegetable pea cultivar ‘Zhewan No. 1’ at the chromosome level and analyzed resequencing data from 314 accessions, creating a comprehensive map of genetic variation in peas. We identified 235 candidate loci associated with 57 important agronomic traits through genome-wide association studies. Notably, we pinpointed the causal gene haplotypes responsible for four Mendelian traits: stem length (Le/le), flower color (A/a), cotyledon color (I/i) and seed shape (R/r). Additionally, we discovered the genes controlling pod form (Mendelian P/p) and hilum color. Our study also involved constructing a gene expression atlas across 22 tissues, highlighting key gene modules related to pod and seed development. These findings provide valuable pea genomic information and will facilitate the future genome-informed improvement of pea crops. Chromosome-scale genome assembly of a vegetable-type elite pea cultivar ZW1 and population analyses using resequencing data of 314 accessions provide insights into the genetic basis of Mendelian and other agronomic traits.
通过对豌豆的参考基因组序列和群体基因组分析,可以深入了解孟德尔性状和其他农艺性状的遗传基础。
豌豆是人类不可或缺的营养品,在孟德尔遗传定律的发现过程中发挥了至关重要的作用。在这项研究中,我们在染色体水平上组装了精英蔬菜豌豆栽培品种 "浙皖 1 号 "的基因组,并分析了来自 314 个登录品系的重测序数据,绘制了一张全面的豌豆遗传变异图谱。通过全基因组关联研究,我们确定了与 57 个重要农艺性状相关的 235 个候选位点。值得注意的是,我们确定了四个孟德尔性状的因果基因单倍型:茎长(Le/le)、花色(A/a)、子叶颜色(I/i)和种子形状(R/r)。此外,我们还发现了控制豆荚形状(孟德尔 P/p)和种脐色的基因。我们的研究还包括构建 22 个组织的基因表达图谱,突出与豆荚和种子发育相关的关键基因模块。这些研究结果提供了宝贵的豌豆基因组信息,将有助于未来对豌豆作物进行基因组信息改良。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature genetics
生物-遗传学
CiteScore
43.00
自引率
2.60%
发文量
241
审稿时长
3 months
期刊介绍:
Nature Genetics publishes the very highest quality research in genetics. It encompasses genetic and functional genomic studies on human and plant traits and on other model organisms. Current emphasis is on the genetic basis for common and complex diseases and on the functional mechanism, architecture and evolution of gene networks, studied by experimental perturbation.
Integrative genetic topics comprise, but are not limited to:
-Genes in the pathology of human disease
-Molecular analysis of simple and complex genetic traits
-Cancer genetics
-Agricultural genomics
-Developmental genetics
-Regulatory variation in gene expression
-Strategies and technologies for extracting function from genomic data
-Pharmacological genomics
-Genome evolution
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


