Geometric deep learning of protein–DNA binding specificity
IF 36.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Predicting protein–DNA binding specificity is a challenging yet essential task for understanding gene regulation. Protein–DNA complexes usually exhibit binding to a selected DNA target site, whereas a protein binds, with varying degrees of binding specificity, to a wide range of DNA sequences. This information is not directly accessible in a single structure. Here, to access this information, we present Deep Predictor of Binding Specificity (DeepPBS), a geometric deep-learning model designed to predict binding specificity from protein–DNA structure. DeepPBS can be applied to experimental or predicted structures. Interpretable protein heavy atom importance scores for interface residues can be extracted. When aggregated at the protein residue level, these scores are validated through mutagenesis experiments. Applied to designed proteins targeting specific DNA sequences, DeepPBS was demonstrated to predict experimentally measured binding specificity. DeepPBS offers a foundation for machine-aided studies that advance our understanding of molecular interactions and guide experimental designs and synthetic biology. DeepPBS is a deep-learning model designed to predict the binding specificity of protein–DNA interactions using physicochemical and geometric contexts. DeepPBS functions across protein families and on experimentally determined as well as predicted protein–DNA complex structures.
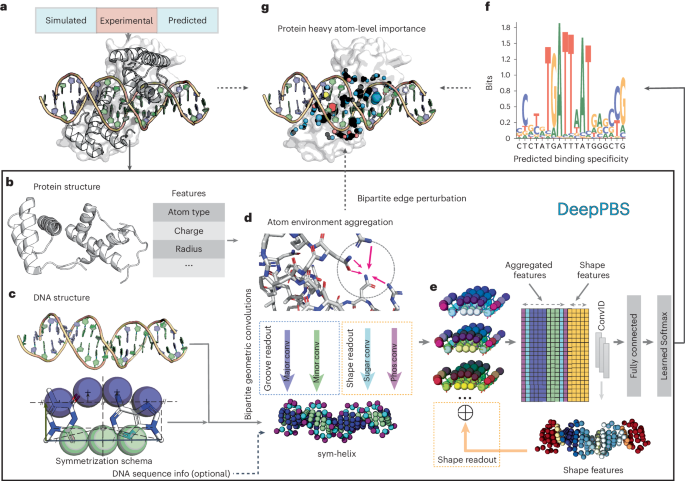
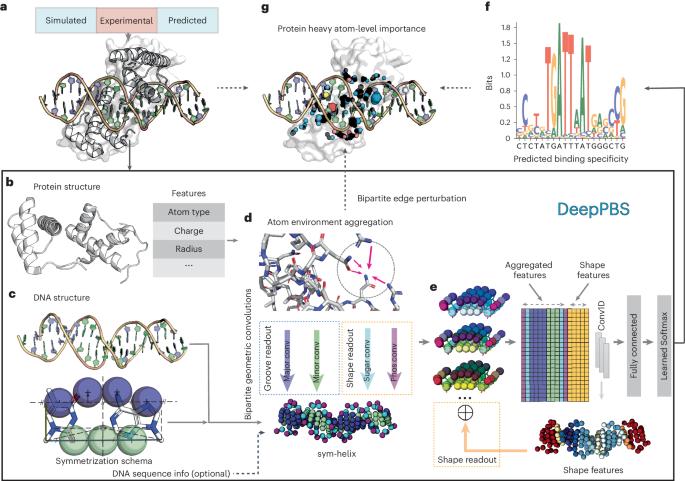
蛋白质-DNA 结合特异性的几何深度学习。
预测蛋白质-DNA 结合的特异性是了解基因调控的一项具有挑战性但又必不可少的任务。蛋白质-DNA 复合物通常表现为与选定的 DNA 目标位点结合,而蛋白质则以不同程度的结合特异性与多种 DNA 序列结合。单个结构无法直接获取这些信息。为了获取这些信息,我们提出了结合特异性深度预测模型(DeepPBS),这是一种几何深度学习模型,旨在从蛋白质-DNA 结构中预测结合特异性。DeepPBS 可应用于实验结构或预测结构。可以为界面残基提取可解释的蛋白质重原子重要性分数。在蛋白质残基水平上汇总后,这些分数可通过诱变实验进行验证。将 DeepPBS 应用于以特定 DNA 序列为靶标的设计蛋白质,证明它可以预测实验测定的结合特异性。DeepPBS 为机器辅助研究奠定了基础,这些研究可促进我们对分子相互作用的理解,并为实验设计和合成生物学提供指导。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Methods
生物-生化研究方法
CiteScore
58.70
自引率
1.70%
发文量
326
审稿时长
1 months
期刊介绍:
Nature Methods is a monthly journal that focuses on publishing innovative methods and substantial enhancements to fundamental life sciences research techniques. Geared towards a diverse, interdisciplinary readership of researchers in academia and industry engaged in laboratory work, the journal offers new tools for research and emphasizes the immediate practical significance of the featured work. It publishes primary research papers and reviews recent technical and methodological advancements, with a particular interest in primary methods papers relevant to the biological and biomedical sciences. This includes methods rooted in chemistry with practical applications for studying biological problems.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


