Systematic loss-of-function screens identify pathway-specific functional circular RNAs
IF 17.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
Circular RNA (circRNA) is covalently closed, single-stranded RNA produced by back-splicing. A few circRNAs have been implicated as functional; however, we lack understanding of pathways that are regulated by circRNAs. Here we generated a pooled short-hairpin RNA library targeting the back-splice junction of 3,354 human circRNAs that are expressed at different levels (ranging from low to high) in humans. We used this library for loss-of-function proliferation screens in a panel of 18 cancer cell lines from four tissue types harbouring mutations leading to constitutive activity of defined pathways. Both context-specific and non-specific circRNAs were identified. Some circRNAs were found to directly regulate their precursor, whereas some have a function unrelated to their precursor. We validated these observations with a secondary screen and uncovered a role for circRERE(4–10) and circHUWE1(22,23), two cell-essential circRNAs, circSMAD2(2–6), a WNT pathway regulator, and circMTO1(2,RI,3), a regulator of MAPK signalling. Our work sheds light on pathways regulated by circRNAs and provides a catalogue of circRNAs with a measurable function. Liu, Neve et al. use large-scale loss-of-function RNA-interference screens to identify circular RNAs that are direct regulators of important signalling pathways and also common essential and tissue-specific circRNAs.
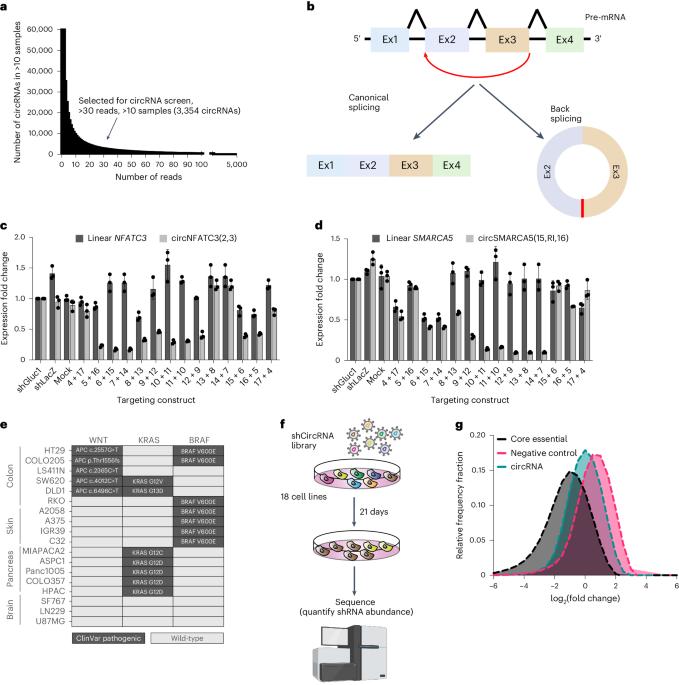
系统性功能缺失筛选确定特异性通路功能环状 RNA
环状 RNA(circRNA)是通过反向剪接产生的共价封闭的单链 RNA。一些 circRNA 被认为具有功能,但我们对 circRNA 的调控途径还缺乏了解。在这里,我们生成了一个以 3,354 个人类 circRNAs 的反向剪接交界处为靶点的短发夹 RNA 文库,这些 circRNAs 在人体内以不同水平(从低到高)表达。我们利用该文库对来自四种组织类型的 18 种癌症细胞系进行了功能缺失增殖筛选,这些细胞系均携带导致特定通路组成性活性的突变。结果发现了特异性和非特异性 circRNA。发现一些 circRNA 可直接调节其前体,而另一些则具有与其前体无关的功能。我们通过二次筛选验证了这些观察结果,并发现了 circRERE(4-10) 和 circHUWE1(22,23)(两种细胞必需的 circRNA)、circSMAD2(2-6)(一种 WNT 通路调控因子)和 circMTO1(2,RI,3)(一种 MAPK 信号调控因子)的作用。我们的工作揭示了受 circRNAs 调节的途径,并提供了具有可测量功能的 circRNAs 目录。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


