expowo: An R package for mining global plant diversity and distribution data
Abstract
Premise
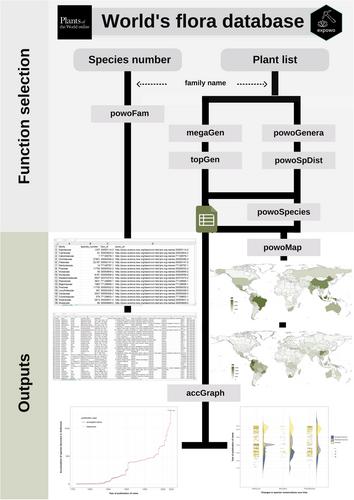
Data on plant distribution and diversity from natural history collections and taxonomic databases are increasingly becoming available online as exemplified by the Royal Botanic Gardens, Kew's Plants of the World Online (POWO) database. This growing accumulation of biodiversity information requires an advance in bioinformatic tools for accessing and processing the massive data for use in downstream science. We present herein expowo, an open-source package that facilitates extracting and using botanical data from POWO.
Methods and Results
The expowo package is implemented in R and designed to handle the entire vascular plant tree of life. It includes functions to readily distill taxonomic and distributional information about all families, genera, or species of vascular plants. It outputs a complete list of species in each genus of any plant family, with the associated original publication, synonyms, and distribution, and plots global maps of species richness at the country and botanical country levels, as well as graphs displaying species-discovery accumulation curves and nomenclatural changes over time. To demonstrate expowo's strengths in producing easy-to-handle outputs, we also show empirical examples from a set of biodiverse countries and representative species-rich and ecologically important angiosperm families.
Conclusions
By harnessing bioinformatic tools that accommodate varying levels of R programming proficiency, expowo functions assist users who have limited R programming expertise in efficiently distilling specific botanical information from online sources and producing maps and graphics for the further interpretation of biogeographic and taxonomic patterns.


 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


