Deep mutational scanning and machine learning for the analysis of antimicrobial-peptide features driving membrane selectivity
IF 26.8
1区 医学
Q1 ENGINEERING, BIOMEDICAL
引用次数: 0
Abstract
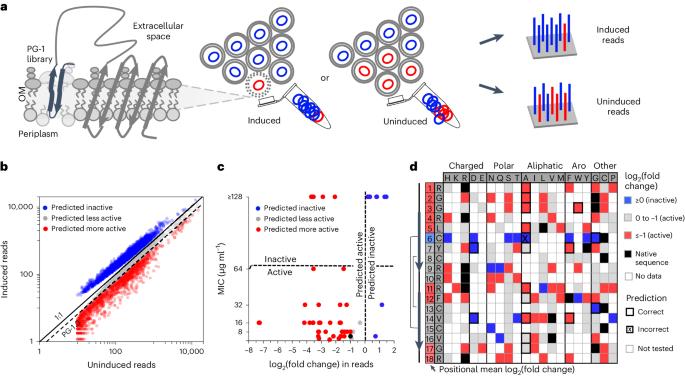
Many antimicrobial peptides directly disrupt bacterial membranes yet can also damage mammalian membranes. It is therefore central to their therapeutic use that rules governing the membrane selectivity of antimicrobial peptides be deciphered. However, this is difficult even for short peptides owing to the large combinatorial space of amino acid sequences. Here we describe a method for measuring the loss or maintenance of antimicrobial-peptide activity for thousands of peptide-sequence variants simultaneously, and its application to Protegrin-1, a potent yet toxic antimicrobial peptide, to determine the positional importance and flexibility of residues across its sequence while identifying variants with changes in membrane selectivity. More bacterially selective variants maintained a membrane-bound secondary structure while avoiding aromatic residues and cysteine pairs. A machine-learning model trained with our datasets accurately predicted membrane-specific activities for over 5.7 million Protegrin-1 variants, and identified one variant that showed substantially reduced toxicity and retention of activity in a mouse model of intraperitoneal infection. The high-throughput methodology may help elucidate sequence–structure–function relationships in antimicrobial peptides and inform the design of peptide-based synthetic drugs. A high-throughput mutation-scanning method for interrogating sequence variation in antimicrobial peptides facilitates the understanding of sequence–structure–function relationships affecting the membrane selectivity of antimicrobial peptides.
利用深度突变扫描和机器学习分析驱动膜选择性的抗菌肽特征
许多抗菌肽能直接破坏细菌膜,但也能破坏哺乳动物膜。因此,破译抗菌肽的膜选择性规则对其治疗用途至关重要。然而,由于氨基酸序列的组合空间很大,即使是短肽也很难做到这一点。在这里,我们介绍了一种同时测量数千个肽序列变体的抗菌肽活性损失或维持情况的方法,并将其应用于 Protegrin-1(一种强效但有毒的抗菌肽),以确定其序列中残基的位置重要性和灵活性,同时识别膜选择性变化的变体。细菌选择性更强的变体保持了膜结合二级结构,同时避免了芳香残基和半胱氨酸对。用我们的数据集训练的机器学习模型准确预测了570多万个Protegrin-1变体的膜特异性活性,并确定了一个变体,它在小鼠腹膜内感染模型中显示出显著的毒性降低和活性保持。这种高通量方法有助于阐明抗菌肽的序列-结构-功能关系,并为设计基于肽的合成药物提供信息。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Biomedical Engineering
Medicine-Medicine (miscellaneous)
CiteScore
45.30
自引率
1.10%
发文量
138
期刊介绍:
Nature Biomedical Engineering is an online-only monthly journal that was launched in January 2017. It aims to publish original research, reviews, and commentary focusing on applied biomedicine and health technology. The journal targets a diverse audience, including life scientists who are involved in developing experimental or computational systems and methods to enhance our understanding of human physiology. It also covers biomedical researchers and engineers who are engaged in designing or optimizing therapies, assays, devices, or procedures for diagnosing or treating diseases. Additionally, clinicians, who make use of research outputs to evaluate patient health or administer therapy in various clinical settings and healthcare contexts, are also part of the target audience.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


