Using neural networks to obtain NMR spectra of both small and macromolecules from blood samples in a single experiment
IF 5.9
2区 化学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
Metabolomics plays a crucial role in understanding metabolic processes within biological systems. Using specific pulse sequences, NMR-based metabolomics detects small and macromolecular metabolites that are altered in blood samples. Here we proposed a method called spectral editing neural network, which can effectively edit and separate the spectral signals of small and macromolecules in 1H NMR spectra of serum and plasma based on the linewidth of the peaks. We applied the model to process the 1H NMR spectra of plasma and serum. The extracted small and macromolecular spectra were then compared with experimentally obtained relaxation-edited and diffusion-edited spectra. Correlation analysis demonstrated the quantitative capability of the model in the extracted small molecule signals from 1H NMR spectra. The principal component analysis showed that the spectra extracted by the model and those obtained by NMR spectral editing methods reveal similar group information, demonstrating the effectiveness of the model in signal extraction. 1H NMR-based metabolomics can detect small and macromolecular metabolites simultaneously from complex biological samples, however, signaling overlap remains a challenge for accurate molecular identification and quantification. Here, the authors develop a spectral editing neural network to effectively edit and separate the spectral signals of small and macromolecules in the 1H NMR spectra of serum and plasma based on the linewidth of the peaks.
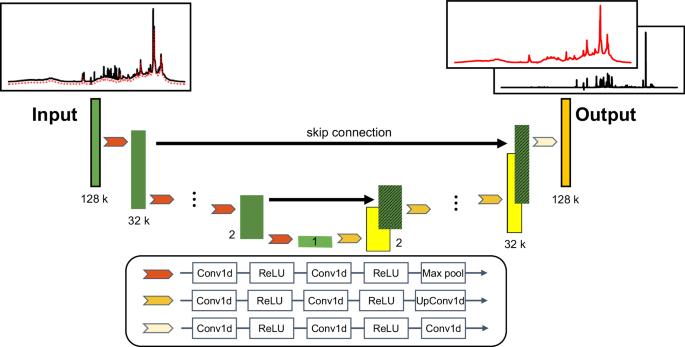
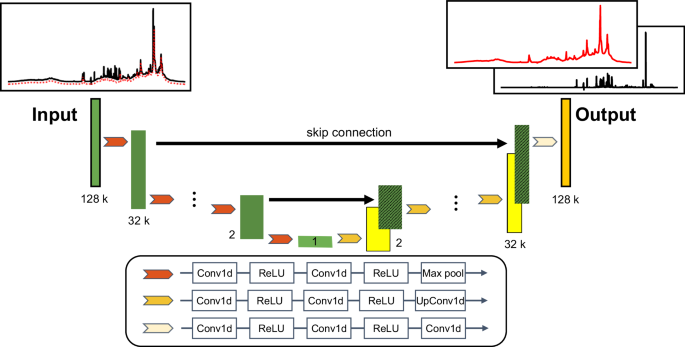
利用神经网络在一次实验中从血液样本中获得小分子和大分子的核磁共振光谱。
代谢组学在了解生物系统内的代谢过程方面发挥着至关重要的作用。利用特定的脉冲序列,基于核磁共振的代谢组学可检测血液样本中发生变化的小分子和大分子代谢物。在此,我们提出了一种称为光谱编辑神经网络的方法,它能根据峰的线宽有效地编辑和分离血清和血浆 1H NMR 光谱中的小分子和大分子光谱信号。我们应用该模型处理了血浆和血清的 1H NMR 光谱。然后将提取的小分子和大分子光谱与实验获得的弛豫编辑光谱和扩散编辑光谱进行比较。相关性分析表明,该模型具有从 1H NMR 图谱中提取小分子信号的定量能力。主成分分析表明,模型提取的光谱与核磁共振光谱编辑方法获得的光谱显示了相似的基团信息,证明了模型在信号提取方面的有效性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Communications Chemistry
Chemistry-General Chemistry
CiteScore
7.70
自引率
1.70%
发文量
146
审稿时长
13 weeks
期刊介绍:
Communications Chemistry is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the chemical sciences. Research papers published by the journal represent significant advances bringing new chemical insight to a specialized area of research. We also aim to provide a community forum for issues of importance to all chemists, regardless of sub-discipline.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


