A network-based trans-omics approach for predicting synergistic drug combinations
IF 5.4
Q1 MEDICINE, RESEARCH & EXPERIMENTAL
引用次数: 0
Abstract
Combination therapy can offer greater efficacy on medical treatments. However, the discovery of synergistic drug combinations is challenging. We propose a novel computational method, SyndrumNET, to predict synergistic drug combinations by network propagation with trans-omics analyses. The prediction is based on the topological relationship, network-based proximity, and transcriptional correlation between diseases and drugs. SyndrumNET was applied to analyzing six diseases including asthma, diabetes, hypertension, colorectal cancer, acute myeloid leukemia (AML), and chronic myeloid leukemia (CML). Here we show that SyndrumNET outperforms the previous methods in terms of high accuracy. We perform in vitro cell survival assays to validate our prediction for CML. Of the top 17 predicted drug pairs, 14 drug pairs successfully exhibits synergistic anticancer effects. Our mode-of-action analysis also reveals that the drug synergy of the top predicted combination of capsaicin and mitoxantrone is due to the complementary regulation of 12 pathways, including the Rap1 signaling pathway. The proposed method is expected to be useful for discovering synergistic drug combinations for various complex diseases. Adding drug treatments together can sometimes produce better results for patients. We introduced a new computer-based method called SyndrumNET, designed to identify effective drug combinations for treating diseases. The method uses data about how diseases and drugs interact at a molecular level to predict which drugs work well together. Tested on six different diseases, such as asthma and different types of cancer, SyndrumNET proved to be more accurate than previous approaches. For example, most of the drug combinations predicted by SyndrumNET to rank highly have shown better combination effects on leukemia cells. This method also helped understand why certain drug combinations work better by analyzing their effects on cellular pathways. The findings suggest that SyndrumNET could be a valuable tool in developing more effective treatment for various complex diseases. Iida et al. predict synergistic drug combinations using a computational method termed SyndrumNET. Validation of predictions in chronic myeloid leukemia using in vitro cell survival assays reveal synergistic anticancer effects in 14 of 17 top predicted drug pairings.
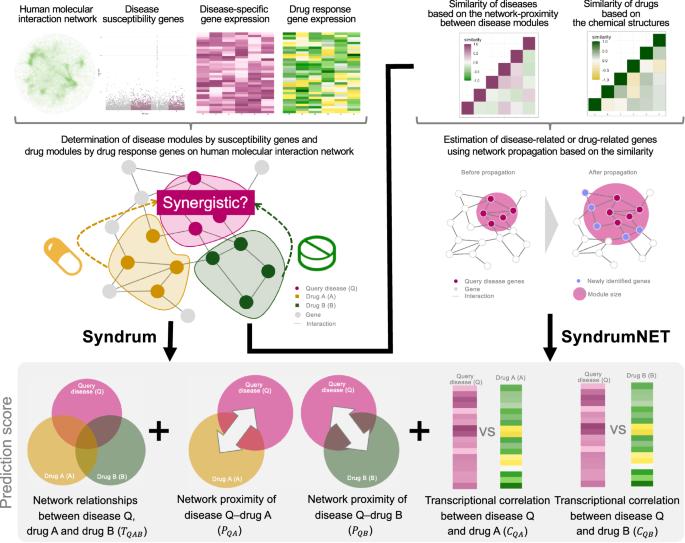
基于网络的跨组学方法预测协同药物组合。
背景介绍联合疗法可以提高医疗效果。然而,发现协同药物组合具有挑战性。我们提出了一种新颖的计算方法--SyndrumNET,通过网络传播和跨组学分析预测协同药物组合:方法:预测基于疾病与药物之间的拓扑关系、基于网络的接近性和转录相关性。SyndrumNET 被应用于分析六种疾病,包括哮喘、糖尿病、高血压、结直肠癌、急性髓性白血病(AML)和慢性髓性白血病(CML):结果:我们在此表明,SyndrumNET 在高精确度方面优于之前的方法。我们进行了体外细胞存活实验来验证我们对 CML 的预测。在排名前 17 位的预测药物配对中,有 14 个药物配对成功显示出协同抗癌效果。我们的作用模式分析还揭示出,辣椒素和米托蒽醌这对最佳预测组合的药物协同作用是由于对包括Rap1信号通路在内的12条通路的互补调控:结论:所提出的方法有望用于发现治疗各种复杂疾病的协同药物组合。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


