Machine-learning and scRNA-Seq-based diagnostic and prognostic models illustrating survival and therapy response of lung adenocarcinoma
IF 4.5
3区 医学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
Lung cancer is a major cause accounting for cancer-related mortalities, with lung adenocarcinoma (LUAD) being the most prevalent subtype. Given the high clinical and cellular heterogeneities of LUAD, accurate diagnosis and prognosis are crucial to avoid overdiagnosis and overtreatment. Taking full advantage of scRNA-Seq data to resolve the tumor heterogeneities, we explored the overall landscape of LUAD microenvironment. Utilizing the stage-specific tumor cell markers, we have developed highly accurate diagnostic and prognostic models with elevated sensitivity and specificity. The diagnostic model, developed through random forest algorithms with a thirteen-gene signature, achieved an accuracy of 96.4% and an AUC of 0.993. These metrics were further demonstrated by benchmarking with available models and scoring systems in independent cohorts. Concurrently, the prognostic model, formulated via Cox regression with a six-gene signature, effectively predicted overall survival, with elevated risk scores associated with increased fractions of cancer-associated fibroblasts, and higher likelihood of immune escape and T-cell exclusion. Subsequently, two nomograms were developed to predict survival and drug responses, facilitating their integration into clinical practice. Overall, this study underscores the potential of our models for efficient, rapid, and cost-effective diagnosis and prognosis of LUAD, adaptable to multiple expression profiling platforms and quantification methods.
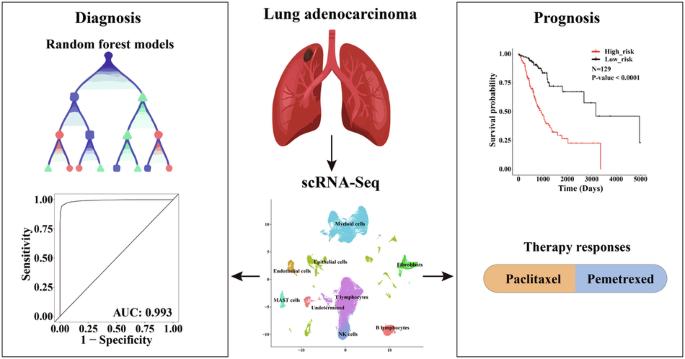
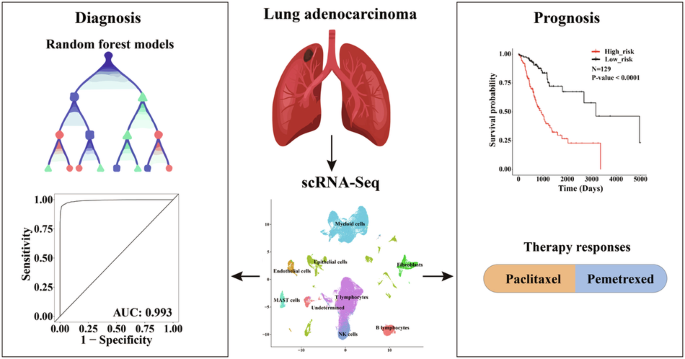
基于机器学习和 scRNA-Seq 的诊断和预后模型,说明肺腺癌的存活率和治疗反应。
肺癌是造成癌症相关死亡的主要原因,其中肺腺癌(LUAD)是最常见的亚型。鉴于肺腺癌的临床和细胞异质性较高,准确诊断和预后对于避免过度诊断和过度治疗至关重要。我们充分利用 scRNA-Seq 数据解决肿瘤异质性问题,探索了 LUAD 微环境的整体情况。利用分期特异性肿瘤细胞标记物,我们开发出了具有较高灵敏度和特异性的高精度诊断和预后模型。通过随机森林算法开发的诊断模型具有 13 个基因特征,准确率达到 96.4%,AUC 为 0.993。通过与独立队列中的现有模型和评分系统进行比对,这些指标得到了进一步证实。同时,通过六基因特征的 Cox 回归建立的预后模型能有效预测总生存期,风险评分的升高与癌症相关成纤维细胞的比例增加以及免疫逃逸和 T 细胞排斥的可能性增加有关。随后,研究人员还开发了两个提名图来预测生存率和药物反应,以便将其应用于临床实践。总之,这项研究强调了我们的模型在高效、快速和经济有效地诊断 LUAD 和预后方面的潜力,并能适应多种表达谱平台和量化方法。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Genes and immunity
医学-免疫学
CiteScore
8.90
自引率
4.00%
发文量
28
审稿时长
6-12 weeks
期刊介绍:
Genes & Immunity emphasizes studies investigating how genetic, genomic and functional variations affect immune cells and the immune system, and associated processes in the regulation of health and disease. It further highlights articles on the transcriptional and posttranslational control of gene products involved in signaling pathways regulating immune cells, and protective and destructive immune responses.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


