Predictions of Chromatography Methods by Chemical Structure Similarity to Accelerate High-Throughput Medicinal Chemistry
IF 3.5
3区 医学
Q2 CHEMISTRY, MEDICINAL
引用次数: 0
Abstract
We introduce a new workflow that relies heavily on chemical quantitative structure-retention relationship (QSRR) models to accelerate method development for micro/mini-scale high-throughput purification (HTP). This provides faster access to new active pharmaceutical ingredients (APIs) through high-throughput experimentation (HTE). By comparing fingerprint structural similarity (e.g., Tanimoto index) with small training data sets containing a few hundred diverse small molecule antagonists of a lipid metabolizing enzyme, we can predict retention time (RT) of new compounds. Machine learning (ML) helps to identify optimal separation conditions for purification without performing the traditional crude QC step involving ultrahigh performance liquid chromatography (UHPLC) analyses of each compound. This green-chemistry approach with the use of predictive tools reduces cost and significantly shortens the design-make-test (DMT) cycle of new drugs by way of HTE.
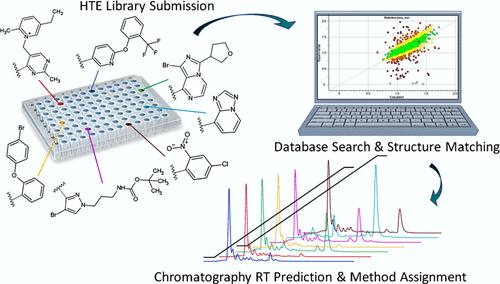
通过化学结构相似性预测色谱方法,加快高通量药物化学的发展
我们介绍了一种新的工作流程,它主要依靠化学定量结构-保留关系(QSRR)模型来加速微/小规模高通量纯化(HTP)的方法开发。这样就能通过高通量实验 (HTE) 更快地获得新的活性药物成分 (API)。通过比较指纹结构相似性(如谷本指数)与包含几百种不同小分子脂质代谢酶拮抗剂的小型训练数据集,我们可以预测新化合物的保留时间(RT)。机器学习(ML)有助于确定纯化的最佳分离条件,而无需执行传统的粗质量控制步骤,即对每个化合物进行超高效液相色谱(UHPLC)分析。这种使用预测工具的绿色化学方法通过 HTE 降低了成本,并大大缩短了新药的设计-制造-测试 (DMT) 周期。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
ACS Medicinal Chemistry Letters
CHEMISTRY, MEDICINAL-
CiteScore
7.30
自引率
2.40%
发文量
328
审稿时长
1 months
期刊介绍:
ACS Medicinal Chemistry Letters is interested in receiving manuscripts that discuss various aspects of medicinal chemistry. The journal will publish studies that pertain to a broad range of subject matter, including compound design and optimization, biological evaluation, drug delivery, imaging agents, and pharmacology of both small and large bioactive molecules. Specific areas include but are not limited to:
Identification, synthesis, and optimization of lead biologically active molecules and drugs (small molecules and biologics)
Biological characterization of new molecular entities in the context of drug discovery
Computational, cheminformatics, and structural studies for the identification or SAR analysis of bioactive molecules, ligands and their targets, etc.
Novel and improved methodologies, including radiation biochemistry, with broad application to medicinal chemistry
Discovery technologies for biologically active molecules from both synthetic and natural (plant and other) sources
Pharmacokinetic/pharmacodynamic studies that address mechanisms underlying drug disposition and response
Pharmacogenetic and pharmacogenomic studies used to enhance drug design and the translation of medicinal chemistry into the clinic
Mechanistic drug metabolism and regulation of metabolic enzyme gene expression
Chemistry patents relevant to the medicinal chemistry field.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


