Mass spectrometry-based proteomic landscape of rice reveals a post-transcriptional regulatory role of N6-methyladenosine
IF 15.8
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
Abstract
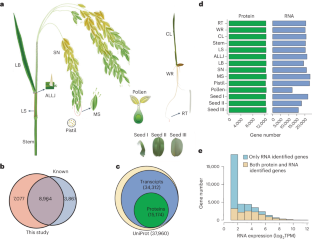
Rice is one of the most important staple food and model species in plant biology, yet its quantitative proteomes are largely uncharacterized. Here we quantify the relative protein levels of over 15,000 genes across major rice tissues using a tandem mass tag strategy followed by intensive fractionation and mass spectrometry. We identify tissue-specific and tissue-enriched proteins that are linked to the functional specificity of individual tissues. Proteogenomic comparison of rice and Arabidopsis reveals conserved proteome expression, which differs from mammals in that there is a strong separation of species rather than tissues. Notably, profiling of N6-methyladenosine (m6A) across the rice major tissues shows that m6A at untranslated regions is negatively correlated with protein abundance and contributes to the discordance between RNA and protein levels. We also demonstrate that our data are valuable for identifying novel genes required for regulating m6A methylation. Taken together, this study provides a paradigm for further research into rice proteogenome. This proteomic landscape study reveals proteins associated with the functional specificity of rice tissues, and further multi-omics analysis shows that N6-methyladenosine in untranslated regions is negatively correlated with protein abundance.

基于质谱的水稻蛋白质组图谱揭示了 N6-甲基腺苷的转录后调控作用
水稻是植物生物学中最重要的主食和模式物种之一,但其定量蛋白质组在很大程度上尚未定性。在这里,我们采用串联质量标签策略,然后进行强化分馏和质谱分析,定量分析了水稻主要组织中超过 15,000 个基因的相对蛋白质水平。我们确定了与各个组织的功能特异性相关的组织特异性蛋白质和组织富集蛋白质。水稻和拟南芥的蛋白质组学比较揭示了蛋白质组表达的保守性,这与哺乳动物的不同之处在于物种而非组织的强烈分离。值得注意的是,对水稻主要组织中的 N6-甲基腺苷(m6A)进行的分析表明,非翻译区的 m6A 与蛋白质丰度呈负相关,是造成 RNA 与蛋白质水平不一致的原因之一。我们还证明,我们的数据对于确定调节 m6A 甲基化所需的新基因很有价值。总之,这项研究为进一步研究水稻蛋白质基因组提供了一个范例。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


