Decoding protein–RNA interactions using CLIP-based methodologies
IF 39.1
1区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
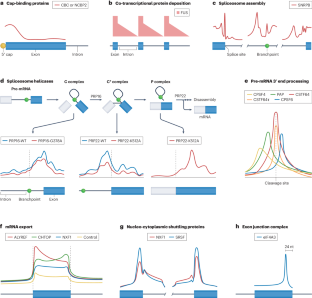
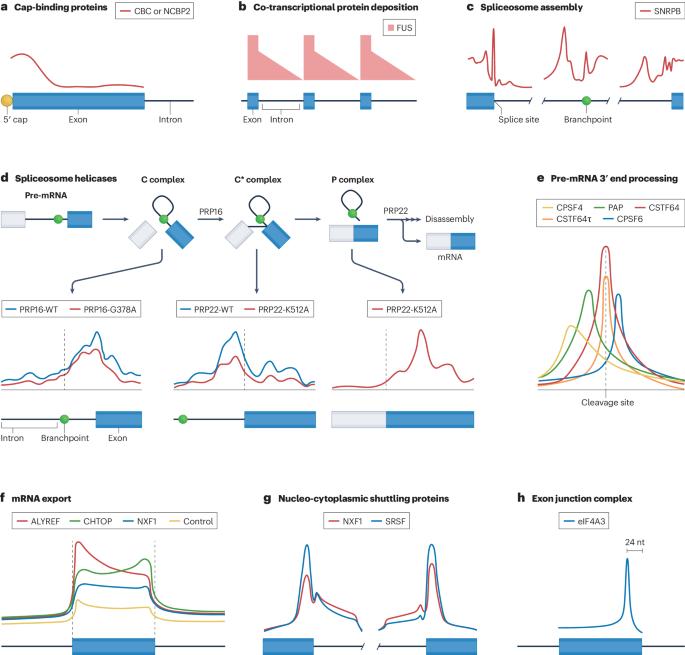
Protein–RNA interactions are central to all RNA processing events, with pivotal roles in the regulation of gene expression and cellular functions. Dysregulation of these interactions has been increasingly linked to the pathogenesis of human diseases. High-throughput approaches to identify RNA-binding proteins and their binding sites on RNA — in particular, ultraviolet crosslinking followed by immunoprecipitation (CLIP) — have helped to map the RNA interactome, yielding transcriptome-wide protein–RNA atlases that have contributed to key mechanistic insights into gene expression and gene-regulatory networks. Here, we review these recent advances, explore the effects of cellular context on RNA binding, and discuss how these insights are shaping our understanding of cellular biology. We also review the potential therapeutic applications arising from new knowledge of protein–RNA interactions. RNA-binding proteins regulate the lifecycle of RNA, and their dysregulation is associated with diseases such as cancer and neurodegeneration. Using methods based on ultraviolet crosslinking followed by immunoprecipitation (CLIP), we can now begin to decode the mechanisms of the interactions between RNA-binding proteins and RNA. This Review discusses recent insights from and future applications of these methodologies.
利用基于 CLIP 的方法解码蛋白质-RNA 之间的相互作用
蛋白质-RNA 相互作用是所有 RNA 处理过程的核心,在调控基因表达和细胞功能方面发挥着关键作用。这些相互作用的失调越来越多地与人类疾病的发病机制联系在一起。鉴定 RNA 结合蛋白及其在 RNA 上结合位点的高通量方法--特别是紫外交联后免疫沉淀(CLIP)--帮助绘制了 RNA 相互作用组图谱,产生了全转录组蛋白质-RNA 图谱,有助于深入了解基因表达和基因调控网络的关键机制。在此,我们回顾了这些最新进展,探讨了细胞环境对 RNA 结合的影响,并讨论了这些见解如何影响我们对细胞生物学的理解。我们还回顾了蛋白质与 RNA 相互作用的新知识可能带来的治疗应用。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Reviews Genetics
生物-遗传学
CiteScore
57.40
自引率
0.50%
发文量
113
审稿时长
6-12 weeks
期刊介绍:
At Nature Reviews Genetics, our goal is to be the leading source of reviews and commentaries for the scientific communities we serve. We are dedicated to publishing authoritative articles that are easily accessible to our readers. We believe in enhancing our articles with clear and understandable figures, tables, and other display items. Our aim is to provide an unparalleled service to authors, referees, and readers, and we are committed to maximizing the usefulness and impact of each article we publish.
Within our journal, we publish a range of content including Research Highlights, Comments, Reviews, and Perspectives that are relevant to geneticists and genomicists. With our broad scope, we ensure that the articles we publish reach the widest possible audience.
As part of the Nature Reviews portfolio of journals, we strive to uphold the high standards and reputation associated with this esteemed collection of publications.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


