A single-cell atlas of chromatin accessibility in mouse organogenesis
IF 17.3
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
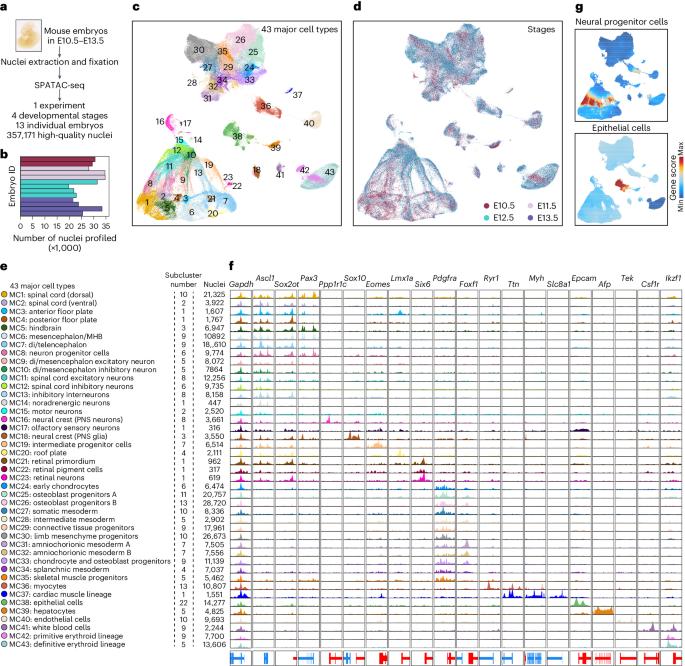
Organogenesis is a highly complex and precisely regulated process. Here we profiled the chromatin accessibility in >350,000 cells derived from 13 mouse embryos at four developmental stages from embryonic day (E) 10.5 to E13.5 by SPATAC-seq in a single experiment. The resulting atlas revealed the status of 830,873 candidate cis-regulatory elements in 43 major cell types. By integrating the chromatin accessibility atlas with the previous transcriptomic dataset, we characterized cis-regulatory sequences and transcription factors associated with cell fate commitment, such as Nr5a2 in the development of gastrointestinal tract, which was preliminarily supported by the in vivo experiment in zebrafish. Finally, we integrated this atlas with the previous single-cell chromatin accessibility dataset from 13 adult mouse tissues to delineate the developmental stage-specific gene regulatory programmes within and across different cell types and identify potential molecular switches throughout lineage development. This comprehensive dataset provides a foundation for exploring transcriptional regulation in organogenesis. In two independent studies, Sun, Liu et al. and Sun et al. develop SPATAC-seq to map the chromatin accessibility landscape of zebrafish embryogenesis and mouse organogenesis, respectively, and identify transcription regulators that determine cell fate.
小鼠器官发生过程中染色质可及性的单细胞图谱
器官发生是一个高度复杂和精确调控的过程。在这里,我们在一次实验中通过 SPATAC-seq 分析了从胚胎 10.5 天到 13.5 天四个发育阶段的 13 个小鼠胚胎的 35 万个细胞的染色质可及性。结果图集揭示了 43 种主要细胞类型中 830,873 个候选顺式调控元件的状态。通过将染色质可及性图谱与之前的转录组数据集整合,我们确定了与细胞命运承诺相关的顺式调控序列和转录因子的特征,如 Nr5a2 在胃肠道发育中的作用,斑马鱼体内实验也初步证实了这一点。最后,我们将这一图集与之前来自 13 个成年小鼠组织的单细胞染色质可及性数据集整合在一起,以勾勒出不同细胞类型内部和跨细胞类型的发育阶段特异性基因调控方案,并识别整个品系发育过程中的潜在分子开关。这个全面的数据集为探索器官发生过程中的转录调控奠定了基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Cell Biology
生物-细胞生物学
CiteScore
28.40
自引率
0.90%
发文量
219
审稿时长
3 months
期刊介绍:
Nature Cell Biology, a prestigious journal, upholds a commitment to publishing papers of the highest quality across all areas of cell biology, with a particular focus on elucidating mechanisms underlying fundamental cell biological processes. The journal's broad scope encompasses various areas of interest, including but not limited to:
-Autophagy
-Cancer biology
-Cell adhesion and migration
-Cell cycle and growth
-Cell death
-Chromatin and epigenetics
-Cytoskeletal dynamics
-Developmental biology
-DNA replication and repair
-Mechanisms of human disease
-Mechanobiology
-Membrane traffic and dynamics
-Metabolism
-Nuclear organization and dynamics
-Organelle biology
-Proteolysis and quality control
-RNA biology
-Signal transduction
-Stem cell biology
文献相关原料
| 公司名称 | 产品信息 | 采购帮参考价格 |
|---|
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


