Zero-shot prediction of mutation effects with multimodal deep representation learning guides protein engineering
IF 28.1
1区 生物学
Q1 CELL BIOLOGY
引用次数: 0
Abstract
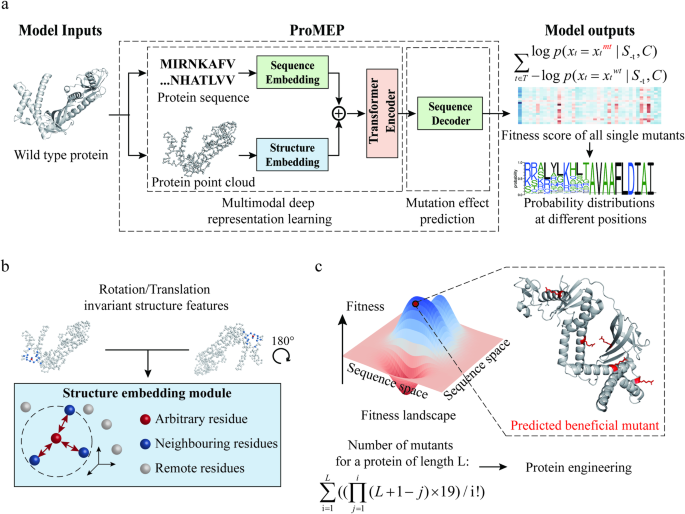
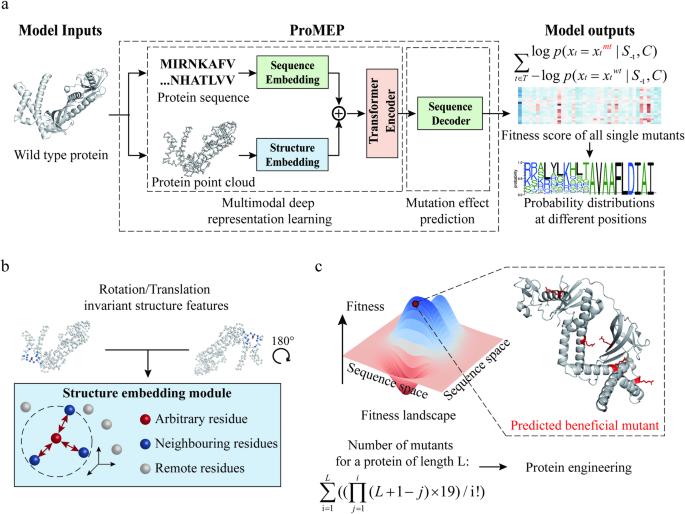
Mutations in amino acid sequences can provoke changes in protein function. Accurate and unsupervised prediction of mutation effects is critical in biotechnology and biomedicine, but remains a fundamental challenge. To resolve this challenge, here we present Protein Mutational Effect Predictor (ProMEP), a general and multiple sequence alignment-free method that enables zero-shot prediction of mutation effects. A multimodal deep representation learning model embedded in ProMEP was developed to comprehensively learn both sequence and structure contexts from ~160 million proteins. ProMEP achieves state-of-the-art performance in mutational effect prediction and accomplishes a tremendous improvement in speed, enabling efficient and intelligent protein engineering. Specifically, ProMEP accurately forecasts mutational consequences on the gene-editing enzymes TnpB and TadA, and successfully guides the development of high-performance gene-editing tools with their engineered variants. The gene-editing efficiency of a 5-site mutant of TnpB reaches up to 74.04% (vs 24.66% for the wild type); and the base editing tool developed on the basis of a TadA 15-site mutant (in addition to the A106V/D108N double mutation that renders deoxyadenosine deaminase activity to TadA) exhibits an A-to-G conversion frequency of up to 77.27% (vs 69.80% for ABE8e, a previous TadA-based adenine base editor) with significantly reduced bystander and off-target effects compared to ABE8e. ProMEP not only showcases superior performance in predicting mutational effects on proteins but also demonstrates a great capability to guide protein engineering. Therefore, ProMEP enables efficient exploration of the gigantic protein space and facilitates practical design of proteins, thereby advancing studies in biomedicine and synthetic biology.
利用多模态深度表征学习零点预测突变效应,为蛋白质工程提供指导。
氨基酸序列的突变会引起蛋白质功能的改变。在生物技术和生物医学中,准确和无监督地预测突变效应至关重要,但这仍然是一个基本挑战。为了解决这一难题,我们在这里提出了蛋白质突变效应预测器(ProMEP),这是一种通用的、无需多序列比对的方法,可以实现突变效应的零次预测。我们开发了一个嵌入 ProMEP 的多模态深度表征学习模型,以从约 1.6 亿个蛋白质中全面学习序列和结构上下文。ProMEP 在突变效应预测方面达到了最先进的性能,并极大地提高了速度,从而实现了高效、智能的蛋白质工程。具体来说,ProMEP 准确预测了基因编辑酶 TnpB 和 TadA 的突变后果,并成功指导了高性能基因编辑工具及其工程变体的开发。TnpB 5 位点突变体的基因编辑效率高达 74.04%(野生型为 24.66%);基于 TadA 15 位点突变体(除了 A106V/D108N 双突变使 TadA 失去脱氧腺苷脱氨酶活性外)开发的碱基编辑工具的 A-G 转换频率高达 77.27%(与之前基于 TadA 的腺嘌呤碱基编辑器 ABE8e 的 69.80% 相比),与 ABE8e 相比,旁观者和脱靶效应显著降低。ProMEP 不仅在预测突变对蛋白质的影响方面表现出卓越的性能,而且在指导蛋白质工程方面也显示出强大的能力。因此,ProMEP 能够有效探索巨大的蛋白质空间,促进蛋白质的实用设计,从而推动生物医学和合成生物学的研究。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cell Research
生物-细胞生物学
CiteScore
53.90
自引率
0.70%
发文量
2420
审稿时长
2.3 months
期刊介绍:
Cell Research (CR) is an international journal published by Springer Nature in partnership with the Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences (CAS). It focuses on publishing original research articles and reviews in various areas of life sciences, particularly those related to molecular and cell biology. The journal covers a broad range of topics including cell growth, differentiation, and apoptosis; signal transduction; stem cell biology and development; chromatin, epigenetics, and transcription; RNA biology; structural and molecular biology; cancer biology and metabolism; immunity and molecular pathogenesis; molecular and cellular neuroscience; plant molecular and cell biology; and omics, system biology, and synthetic biology. CR is recognized as China's best international journal in life sciences and is part of Springer Nature's prestigious family of Molecular Cell Biology journals.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


