Geometry-complete diffusion for 3D molecule generation and optimization
IF 5.9
2区 化学
Q1 CHEMISTRY, MULTIDISCIPLINARY
引用次数: 0
Abstract
Generative deep learning methods have recently been proposed for generating 3D molecules using equivariant graph neural networks (GNNs) within a denoising diffusion framework. However, such methods are unable to learn important geometric properties of 3D molecules, as they adopt molecule-agnostic and non-geometric GNNs as their 3D graph denoising networks, which notably hinders their ability to generate valid large 3D molecules. In this work, we address these gaps by introducing the Geometry-Complete Diffusion Model (GCDM) for 3D molecule generation, which outperforms existing 3D molecular diffusion models by significant margins across conditional and unconditional settings for the QM9 dataset and the larger GEOM-Drugs dataset, respectively. Importantly, we demonstrate that GCDM’s generative denoising process enables the model to generate a significant proportion of valid and energetically-stable large molecules at the scale of GEOM-Drugs, whereas previous methods fail to do so with the features they learn. Additionally, we show that extensions of GCDM can not only effectively design 3D molecules for specific protein pockets but can be repurposed to consistently optimize the geometry and chemical composition of existing 3D molecules for molecular stability and property specificity, demonstrating new versatility of molecular diffusion models. Code and data are freely available on GitHub . Geometric deep learning methods have the advantage of being expressive, while denoising diffusion probabilistic models have great generative power. Here, the authors introduce a geometry-complete diffusion model for effective 3D molecule generation for specific protein pockets that can also consistently optimize the geometry and chemical composition of existing 3D molecules for molecular stability and property specificity
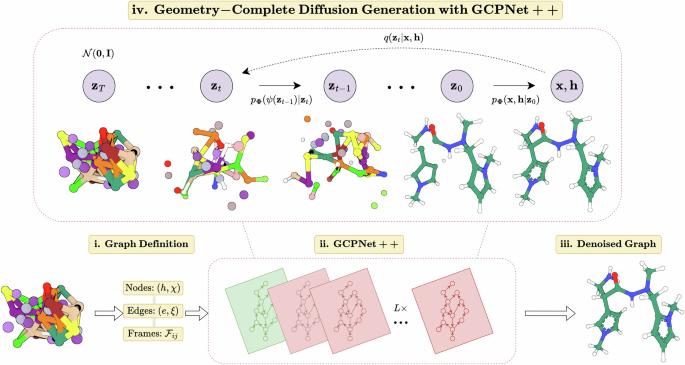
用于三维分子生成和优化的几何完全扩散。
最近有人提出了在去噪扩散框架内使用等变图神经网络(GNN)生成三维分子的生成性深度学习方法。然而,这些方法无法学习三维分子的重要几何特性,因为它们采用了与分子无关的非几何 GNN 作为三维图去噪网络,这明显阻碍了它们生成有效大型三维分子的能力。在这项工作中,我们引入了用于生成三维分子的几何完全扩散模型(GCDM),从而弥补了这些不足。在 QM9 数据集和更大的 GEOM-Drugs 数据集上,GCDM 在有条件和无条件设置下分别以显著的优势优于现有的三维分子扩散模型。重要的是,我们证明了 GCDM 的生成去噪过程能使模型在 GEOM-Drugs 的尺度上生成相当大比例的有效且能量稳定的大分子,而之前的方法无法利用其学习到的特征做到这一点。此外,我们还表明,GCDM 的扩展不仅可以有效地为特定的蛋白质口袋设计三维分子,还可以重新利用它来持续优化现有三维分子的几何形状和化学成分,以实现分子稳定性和特性特异性,从而展示了分子扩散模型新的多功能性。代码和数据可在 GitHub 上免费获取。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Communications Chemistry
Chemistry-General Chemistry
CiteScore
7.70
自引率
1.70%
发文量
146
审稿时长
13 weeks
期刊介绍:
Communications Chemistry is an open access journal from Nature Research publishing high-quality research, reviews and commentary in all areas of the chemical sciences. Research papers published by the journal represent significant advances bringing new chemical insight to a specialized area of research. We also aim to provide a community forum for issues of importance to all chemists, regardless of sub-discipline.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


