Temporal dynamics and persistence of resistance genes to broad spectrum antibiotics in an urban community
IF 10.4
1区 工程技术
Q1 ENGINEERING, CHEMICAL
引用次数: 0
Abstract
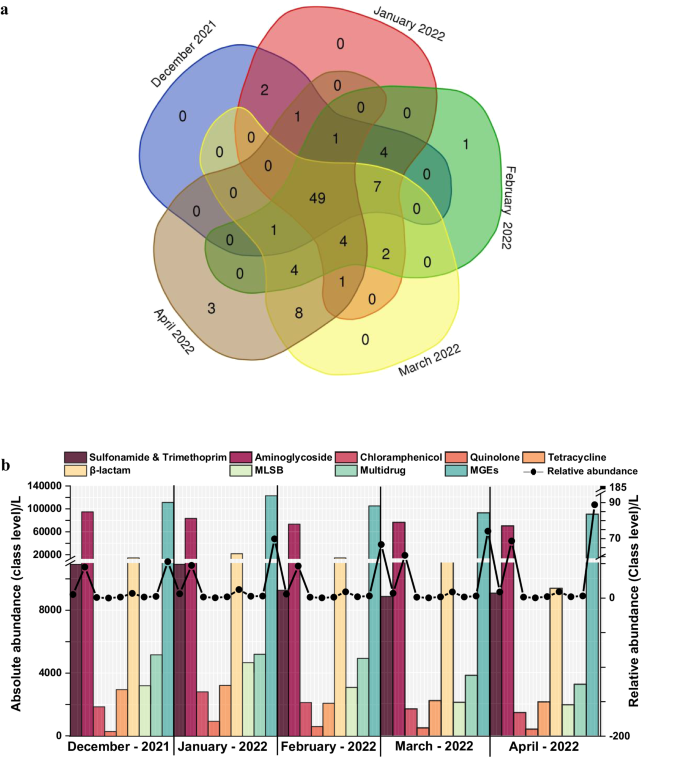
The constantly evolving and growing global health crisis, antimicrobial resistance (AMR), endangers progress in medicine, food production, and life expectancy. Limited data on population-level prevalence, including seasonal and temporal variations, hampers accurate risk assessment for AMR transmission, despite its paramount importance on both global and national fronts. In this study, we used quantitative real-time PCR (q-PCR) to analyze 123 antibiotic resistance genes (ARGs) and 13 mobile genetic elements (MGEs) in wastewater of a selected urban community. Sampling was conducted monthly over a 5-month period (December 2021–April 2022) to assess resistance diversity, temporal dynamics, co-abundance of ARGs, MGEs, and resistance mechanisms. Approximately 50% of the tested ARG subtypes were consistently detected in all months, with frequency ranging from 52 to 61% with maximum absolute abundance in the winter months (December and January). In co-abundance analysis, specific genes were clustered into modules, highlighting shared distribution patterns and functional associations among the ARGs and MGEs. Detected clinically significant genes (ndm-1 and cfiA) and other variants (blaoxy, aph, aacC, tet-35, tet M, tet-32) are capable of imparting resistance to 3rd and 4th generation (gen) β-lactam, aminoglycoside, tetracycline, and multidrug classes. These contribute significantly to core/persistent resistance. This study deepens our comprehension of temporal/seasonal fluctuations in ARG and MGE distribution, providing valuable evidence to guide AMR control policies and promote responsible antibiotic/antimicrobial use for preserving effectiveness.
城市社区广谱抗生素耐药基因的时间动态和持久性
抗菌素耐药性(AMR)是一个不断演变和增长的全球健康危机,它危及医药、食品生产和预期寿命的进步。尽管 AMR 对全球和国家都至关重要,但包括季节性和时间性变化在内的人群水平流行率数据有限,阻碍了对 AMR 传播风险的准确评估。在这项研究中,我们使用定量实时 PCR(q-PCR)技术分析了选定城市社区废水中的 123 个抗生素耐药基因 (ARG) 和 13 个移动遗传因子 (MGE)。在为期 5 个月(2021 年 12 月至 2022 年 4 月)的时间里,每月进行一次采样,以评估耐药性多样性、时间动态、ARGs 和 MGEs 的共丰度以及耐药性机制。在所有月份中都能持续检测到约 50%的受测 ARG 亚型,检测频率在 52% 到 61% 之间,冬季月份(12 月和 1 月)的绝对丰度最高。在共丰度分析中,特定基因被聚类为模块,突出了ARGs和MGEs之间的共同分布模式和功能关联。检测到的具有临床意义的基因(ndm-1 和 cfiA)和其他变体(blaoxy、aph、acC、tet-35、tet M、tet-32)能够产生对第三代和第四代(基因)β-内酰胺类、氨基糖苷类、四环素类和多种药物的耐药性。这些因素在很大程度上导致了核心/持久耐药性。这项研究加深了我们对 ARG 和 MGE 分布的时间/季节波动的理解,为指导 AMR 控制政策和促进负责任地使用抗生素/抗菌药以保持有效性提供了宝贵的证据。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

npj Clean Water
Environmental Science-Water Science and Technology
CiteScore
15.30
自引率
2.60%
发文量
61
审稿时长
5 weeks
期刊介绍:
npj Clean Water publishes high-quality papers that report cutting-edge science, technology, applications, policies, and societal issues contributing to a more sustainable supply of clean water. The journal's publications may also support and accelerate the achievement of Sustainable Development Goal 6, which focuses on clean water and sanitation.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


