Pan-cancer profiling of tumor-infiltrating natural killer cells through transcriptional reference mapping
IF 27.7
1区 医学
Q1 IMMUNOLOGY
引用次数: 0
Abstract
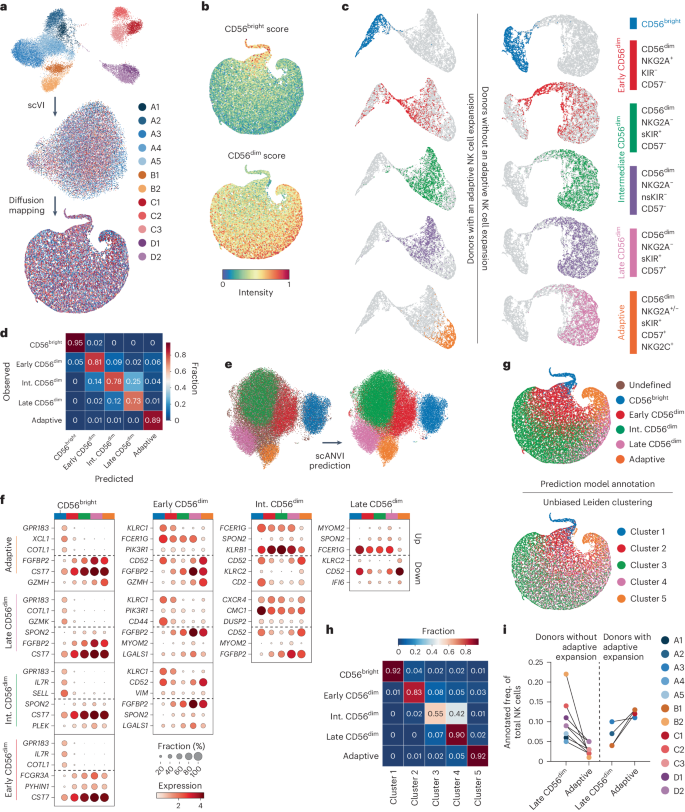
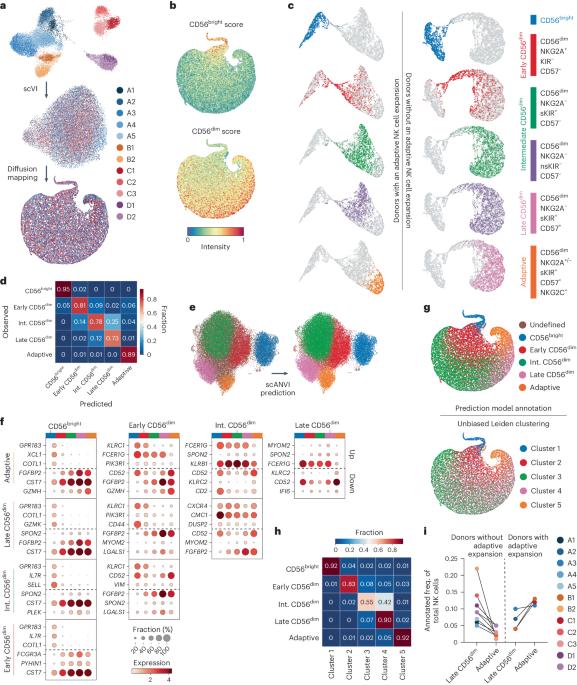
The functional diversity of natural killer (NK) cell repertoires stems from differentiation, homeostatic, receptor–ligand interactions and adaptive-like responses to viral infections. In the present study, we generated a single-cell transcriptional reference map of healthy human blood- and tissue-derived NK cells, with temporal resolution and fate-specific expression of gene-regulatory networks defining NK cell differentiation. Transfer learning facilitated incorporation of tumor-infiltrating NK cell transcriptomes (39 datasets, 7 solid tumors, 427 patients) into the reference map to analyze tumor microenvironment (TME)-induced perturbations. Of the six functionally distinct NK cell states identified, a dysfunctional stressed CD56bright state susceptible to TME-induced immunosuppression and a cytotoxic TME-resistant effector CD56dim state were commonly enriched across tumor types, the ratio of which was predictive of patient outcome in malignant melanoma and osteosarcoma. This resource may inform the design of new NK cell therapies and can be extended through transfer learning to interrogate new datasets from experimental perturbations or disease conditions. Malmberg and colleagues generated a single-cell transcriptional reference map to investigate pan-cancer profiles of tumor-infiltrating natural killer cells.
通过转录参考图谱绘制肿瘤浸润自然杀伤细胞的泛癌症图谱
自然杀伤细胞(NK)的功能多样性源于分化、同源性、受体配体相互作用以及对病毒感染的适应性反应。在本研究中,我们生成了健康人类血液和组织来源NK细胞的单细胞转录参考图谱,该图谱具有时间分辨率和定义NK细胞分化的基因调控网络的命运特异性表达。迁移学习有助于将肿瘤浸润NK细胞转录组(39个数据集,7种实体瘤,427名患者)纳入参考图谱,以分析肿瘤微环境(TME)诱导的扰动。在鉴定出的六种功能不同的NK细胞状态中,易受TME诱导的免疫抑制的功能失调受压CD56bright状态和细胞毒性TME抗性效应CD56dim状态在不同肿瘤类型中普遍富集,两者的比例可预测恶性黑色素瘤和骨肉瘤患者的预后。这一资源可为设计新的NK细胞疗法提供信息,并可通过迁移学习进行扩展,以分析来自实验扰动或疾病状况的新数据集。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Immunology
医学-免疫学
CiteScore
40.00
自引率
2.30%
发文量
248
审稿时长
4-8 weeks
期刊介绍:
Nature Immunology is a monthly journal that publishes the highest quality research in all areas of immunology. The editorial decisions are made by a team of full-time professional editors. The journal prioritizes work that provides translational and/or fundamental insight into the workings of the immune system. It covers a wide range of topics including innate immunity and inflammation, development, immune receptors, signaling and apoptosis, antigen presentation, gene regulation and recombination, cellular and systemic immunity, vaccines, immune tolerance, autoimmunity, tumor immunology, and microbial immunopathology. In addition to publishing significant original research, Nature Immunology also includes comments, News and Views, research highlights, matters arising from readers, and reviews of the literature. The journal serves as a major conduit of top-quality information for the immunology community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


