Applications of artificial intelligence in the analysis of histopathology images of gliomas: a review
引用次数: 0
Abstract
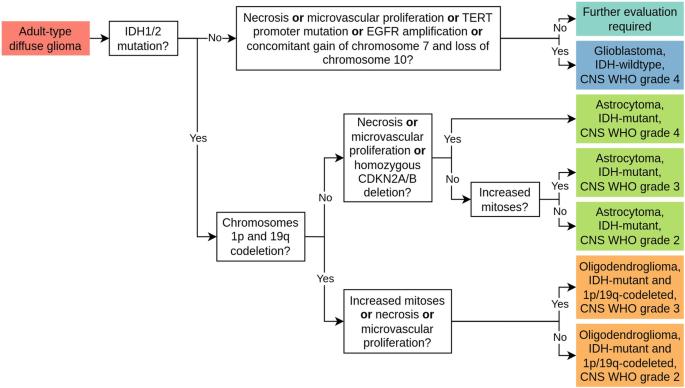
In recent years, the diagnosis of gliomas has become increasingly complex. Analysis of glioma histopathology images using artificial intelligence (AI) offers new opportunities to support diagnosis and outcome prediction. To give an overview of the current state of research, this review examines 83 publicly available research studies that have proposed AI-based methods for whole-slide histopathology images of human gliomas, covering the diagnostic tasks of subtyping (23/83), grading (27/83), molecular marker prediction (20/83), and survival prediction (29/83). All studies were reviewed with regard to methodological aspects as well as clinical applicability. It was found that the focus of current research is the assessment of hematoxylin and eosin-stained tissue sections of adult-type diffuse gliomas. The majority of studies (52/83) are based on the publicly available glioblastoma and low-grade glioma datasets from The Cancer Genome Atlas (TCGA) and only a few studies employed other datasets in isolation (16/83) or in addition to the TCGA datasets (15/83). Current approaches mostly rely on convolutional neural networks (63/83) for analyzing tissue at 20x magnification (35/83). A new field of research is the integration of clinical data, omics data, or magnetic resonance imaging (29/83). So far, AI-based methods have achieved promising results, but are not yet used in real clinical settings. Future work should focus on the independent validation of methods on larger, multi-site datasets with high-quality and up-to-date clinical and molecular pathology annotations to demonstrate routine applicability.
人工智能在胶质瘤组织病理学图像分析中的应用:综述
近年来,胶质瘤的诊断变得越来越复杂。利用人工智能(AI)分析胶质瘤组织病理学图像为支持诊断和结果预测提供了新的机遇。为了概述目前的研究状况,本综述对 83 项公开发表的研究进行了审查,这些研究提出了基于人工智能的人类胶质瘤全切片组织病理学图像分析方法,涵盖了亚型分类(23/83)、分级(27/83)、分子标记预测(20/83)和生存预测(29/83)等诊断任务。对所有研究的方法学方面和临床适用性进行了审查。研究发现,目前研究的重点是对成人型弥漫性胶质瘤苏木精和伊红染色的组织切片进行评估。大多数研究(52/83)都是基于癌症基因组图谱(TCGA)中公开的胶质母细胞瘤和低级别胶质瘤数据集,只有少数研究单独(16/83)或在TCGA数据集之外(15/83)使用了其他数据集。目前的方法大多依赖卷积神经网络(63/83)来分析放大 20 倍的组织(35/83)。一个新的研究领域是整合临床数据、omics 数据或磁共振成像(29/83)。迄今为止,基于人工智能的方法已经取得了可喜的成果,但尚未用于实际临床环境。未来的工作应侧重于在具有高质量和最新临床及分子病理学注释的大型多站点数据集上对方法进行独立验证,以证明其常规适用性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


