CohortFinder: an open-source tool for data-driven partitioning of digital pathology and imaging cohorts to yield robust machine-learning models
引用次数: 0
Abstract
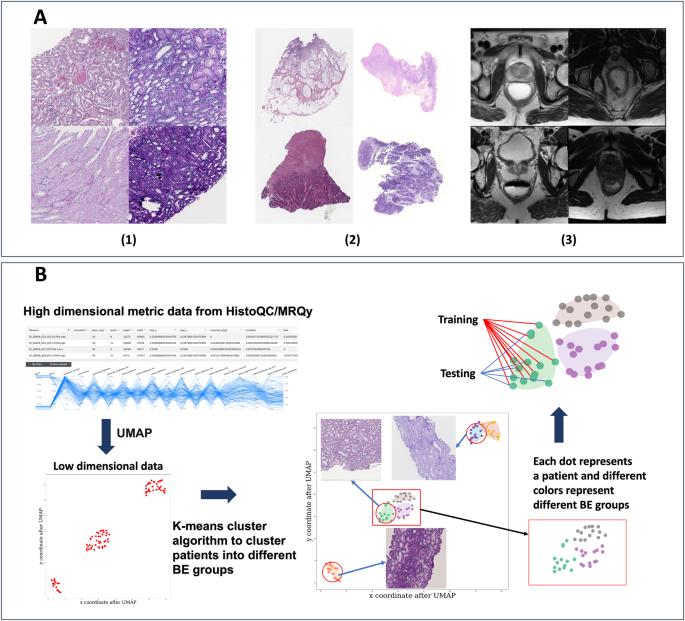
Batch effects (BEs) refer to systematic technical differences in data collection unrelated to biological variations whose noise is shown to negatively impact machine learning (ML) model generalizability. Here we release CohortFinder ( http://cohortfinder.com ), an open-source tool aimed at mitigating BEs via data-driven cohort partitioning. We demonstrate CohortFinder improves ML model performance in downstream digital pathology and medical image processing tasks. CohortFinder is freely available for download at cohortfinder.com.
CohortFinder:一种开源工具,用于对数字病理学和成像队列进行数据驱动的分区,以建立强大的机器学习模型
批次效应(BEs)指的是数据收集中与生物变异无关的系统性技术差异,其噪声已被证明会对机器学习(ML)模型的普适性产生负面影响。在此,我们发布了 CohortFinder ( http://cohortfinder.com ),这是一款开源工具,旨在通过数据驱动的队列分区来减轻批次效应。我们展示了 CohortFinder 在下游数字病理学和医学图像处理任务中提高了 ML 模型性能。CohortFinder 可在 cohortfinder.com 上免费下载。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


