Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
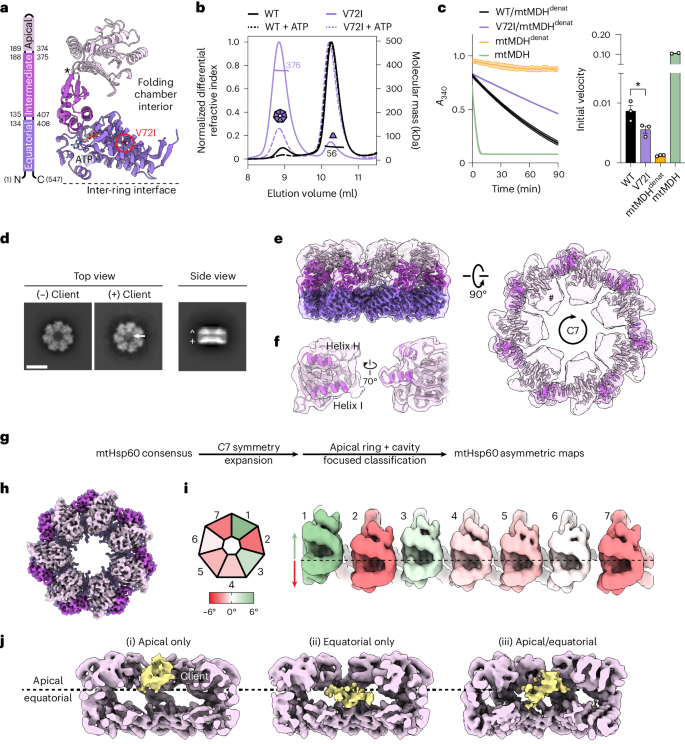
The mitochondrial chaperonin, mitochondrial heat shock protein 60 (mtHsp60), promotes the folding of newly imported and transiently misfolded proteins in the mitochondrial matrix, assisted by its co-chaperone mtHsp10. Despite its essential role in mitochondrial proteostasis, structural insights into how this chaperonin progresses through its ATP-dependent client folding cycle are not clear. Here, we determined cryo-EM structures of a hyperstable disease-associated human mtHsp60 mutant, V72I. Client density is identified in three distinct states, revealing interactions with the mtHsp60 apical domains and C termini that coordinate client positioning in the folding chamber. We further identify an asymmetric arrangement of the apical domains in the ATP state, in which an alternating up/down configuration positions interaction surfaces for simultaneous recruitment of mtHsp10 and client retention. Client is then fully encapsulated in mtHsp60–10, revealing prominent contacts at two discrete sites that potentially support maturation. These results identify distinct roles for the apical domains in coordinating client capture and progression through the chaperone cycle, supporting a conserved mechanism of group I chaperonin function. Cryo-EM structures of the human mitochondrial chaperone Hsp60 reveal alternating apical domain conformations that enable simultaneous client and co-chaperone interactions. These results provide a structural mechanism for efficient client capture and chaperone cycling.

线粒体 Hsp60 的非对称顶端结构域状态协调底物啮合和伴侣蛋白组装
线粒体合子蛋白--线粒体热休克蛋白 60(mtHsp60)在其辅助合子 mtHsp10 的协助下,促进线粒体基质中新导入蛋白质和瞬时折叠错误蛋白质的折叠。尽管它在线粒体蛋白稳态中发挥着重要作用,但人们对这种伴侣蛋白如何通过其依赖 ATP 的客户折叠周期进行折叠的结构研究并不清楚。在这里,我们测定了与疾病相关的人类 mtHsp60 超稳定突变体 V72I 的冷冻电镜结构。在三种不同的状态下确定了客户密度,揭示了与 mtHsp60 顶端结构域和 C 末端的相互作用,这些相互作用协调了客户在折叠室中的定位。我们进一步确定了顶端结构域在 ATP 状态下的不对称排列,在这种状态下,上下交替的配置定位了相互作用表面,以便同时招募 mtHsp10 和保留客户。然后,客户被完全包裹在 mtHsp60-10 中,显示出两个离散位点的突出接触,这两个位点可能支持成熟。这些结果确定了顶端结构域在协调客户捕获和伴侣循环过程中的不同作用,支持了 I 组伴侣素功能的保守机制。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


