Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
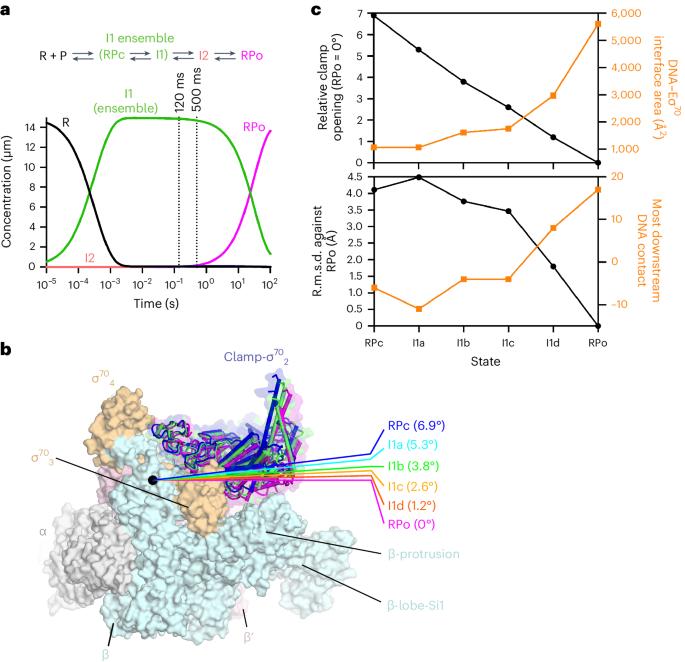
During formation of the transcription-competent open complex (RPo) by bacterial RNA polymerases (RNAPs), transient intermediates pile up before overcoming a rate-limiting step. Structural descriptions of these interconversions in real time are unavailable. To address this gap, here we use time-resolved cryogenic electron microscopy (cryo-EM) to capture four intermediates populated 120 ms or 500 ms after mixing Escherichia coli σ70–RNAP and the λPR promoter. Cryo-EM snapshots revealed that the upstream edge of the transcription bubble unpairs rapidly, followed by stepwise insertion of two conserved nontemplate strand (nt-strand) bases into RNAP pockets. As the nt-strand ‘read-out’ extends, the RNAP clamp closes, expelling an inhibitory σ70 domain from the active-site cleft. The template strand is fully unpaired by 120 ms but remains dynamic, indicating that yet unknown conformational changes complete RPo formation in subsequent steps. Given that these events likely describe DNA opening at many bacterial promoters, this study provides insights into how DNA sequence regulates steps of RPo formation. Time-resolved cryo-EM captured transient intermediates during E. coli RNAP promoter melting, revealing conformational changes affecting stepwise transcription bubble opening. Results inform how DNA sequence controls bacterial transcription initiation.
用时间分辨冷冻电镜观察细菌 RNA 聚合酶启动子熔化的早期中间产物
在细菌 RNA 聚合酶(RNAPs)形成转录功能开放复合物(RPo)的过程中,瞬时中间产物在克服限速步骤之前会堆积起来。目前还没有关于这些相互转化的实时结构描述。为了填补这一空白,我们在此使用时间分辨低温电子显微镜(cryo-EM)捕捉大肠杆菌σ70-RNAP与λPR启动子混合后120毫秒或500毫秒内产生的四个中间产物。低温电子显微镜快照显示,转录泡的上游边缘迅速解除配对,随后两个保守的非模板链(nt-strand)碱基逐步插入 RNAP 口袋。随着 nt 链 "读出 "的延伸,RNAP 夹闭,将抑制性 σ70 结构域从活性位点裂隙中排出。模板链在 120 毫秒前完全未配对,但仍保持动态,这表明在随后的步骤中,未知的构象变化完成了 RPo 的形成。鉴于这些事件很可能描述了许多细菌启动子的 DNA 开启过程,本研究提供了有关 DNA 序列如何调控 RPo 形成步骤的见解。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


