A review on the diversity of antimicrobial peptides and genome mining strategies for their prediction
IF 3.3
3区 生物学
Q2 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
Antibiotic resistance has become one of the most serious threats to human health in recent years. In response to the increasing microbial resistance to the antibiotics currently available, it is imperative to develop new antibiotics or explore new approaches to combat antibiotic resistance. Antimicrobial peptides (AMPs) have shown considerable promise in this regard, as the microbes develop low or no resistance against them. The discovery and development of AMPs still confront numerous obstacles such as finding a target, developing assays, and identifying hits and leads, which are time-consuming processes, making it difficult to reach the market. However, with the advent of genome mining, new antibiotics could be discovered efficiently using tools such as BAGEL, antiSMASH, RODEO, etc., providing hope for better treatment of diseases in the future. Computational methods used in genome mining automatically detect and annotate biosynthetic gene clusters in genomic data, making it a useful tool in natural product discovery. This review aims to shed light on the history, diversity, and mechanisms of action of AMPs and the data on new AMPs identified by traditional as well as genome mining strategies. It further substantiates the various phases of clinical trials for some AMPs, as well as an overview of genome mining databases and tools built expressly for AMP discovery. In light of the recent advancements, it is evident that targeted genome mining stands as a beacon of hope, offering immense potential to expedite the discovery of novel antimicrobials.

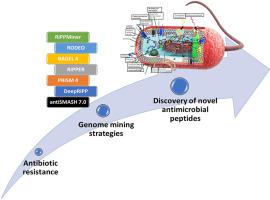
抗菌肽多样性及其预测基因组挖掘策略综述。
近年来,抗生素耐药性已成为人类健康面临的最严重威胁之一。为了应对微生物对现有抗生素日益增长的耐药性,开发新的抗生素或探索新的方法来对抗抗生素耐药性势在必行。在这方面,抗菌肽(AMPs)已显示出相当大的前景,因为微生物对其产生的抗药性很低或没有抗药性。AMPs 的发现和开发仍面临许多障碍,如寻找靶点、开发检测方法、确定命中和线索等,这些都是耗时的过程,因此很难进入市场。然而,随着基因组挖掘技术的出现,人们可以利用 BAGEL、antiSMASH、RODEO 等工具高效地发现新的抗生素,为未来更好地治疗疾病带来希望。基因组挖掘中使用的计算方法能自动检测和注释基因组数据中的生物合成基因簇,使其成为天然产物发现的有用工具。本综述旨在阐明 AMPs 的历史、多样性和作用机制,以及通过传统和基因组挖掘策略发现的新 AMPs 数据。它进一步证实了一些 AMPs 临床试验的各个阶段,并概述了专门为发现 AMP 而建立的基因组挖掘数据库和工具。鉴于最近取得的进展,有针对性的基因组挖掘显然是一盏希望的明灯,为加快新型抗菌药物的发现提供了巨大的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Biochimie
生物-生化与分子生物学
CiteScore
7.20
自引率
2.60%
发文量
219
审稿时长
40 days
期刊介绍:
Biochimie publishes original research articles, short communications, review articles, graphical reviews, mini-reviews, and hypotheses in the broad areas of biology, including biochemistry, enzymology, molecular and cell biology, metabolic regulation, genetics, immunology, microbiology, structural biology, genomics, proteomics, and molecular mechanisms of disease. Biochimie publishes exclusively in English.
Articles are subject to peer review, and must satisfy the requirements of originality, high scientific integrity and general interest to a broad range of readers. Submissions that are judged to be of sound scientific and technical quality but do not fully satisfy the requirements for publication in Biochimie may benefit from a transfer service to a more suitable journal within the same subject area.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


