Spatially resolved epigenome sequencing via Tn5 transposition and deterministic DNA barcoding in tissue
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Spatial epigenetic mapping of tissues enables the study of gene regulation programs and cellular functions with the dependency on their local tissue environment. Here we outline a complete procedure for two spatial epigenomic profiling methods: spatially resolved genome-wide profiling of histone modifications using in situ cleavage under targets and tagmentation (CUT&Tag) chemistry (spatial-CUT&Tag) and transposase-accessible chromatin sequencing (spatial-ATAC-sequencing) for chromatin accessibility. Both assays utilize in-tissue Tn5 transposition to recognize genomic DNA loci followed by microfluidic deterministic barcoding to incorporate spatial address codes. Furthermore, these two methods do not necessitate prior knowledge of the transcription or epigenetic markers for a given tissue or cell type but permit genome-wide unbiased profiling pixel-by-pixel at the 10 μm pixel size level and single-base resolution. To support the widespread adaptation of these methods, details are provided in five general steps: (1) sample preparation; (2) Tn5 transposition in spatial-ATAC-sequencing or antibody-controlled pA–Tn5 tagmentation in CUT&Tag; (3) library preparation; (4) next-generation sequencing; and (5) data analysis using our customed pipelines available at: https://github.com/dyxmvp/Spatial_ATAC-seq and https://github.com/dyxmvp/spatial-CUT-Tag . The whole procedure can be completed on four samples in 2–3 days. Familiarity with basic molecular biology and bioinformatics skills with access to a high-performance computing environment are required. A rudimentary understanding of pathology and specimen sectioning, as well as deterministic barcoding in tissue-specific skills (e.g., design of a multiparameter barcode panel and creation of microfluidic devices), are also advantageous. In this protocol, we mainly focus on spatial profiling of tissue region-specific epigenetic landscapes in mouse embryos and mouse brains using spatial-ATAC-sequencing and spatial-CUT&Tag, but these methods can be used for other species with no need for species-specific probe design. Deterministic barcoding in tissue allows the mapping of chromatin accessibility and histone modifications with high spatial resolution via next-generation sequencing. The method enables rapid identification of cell types and their spatial distribution.
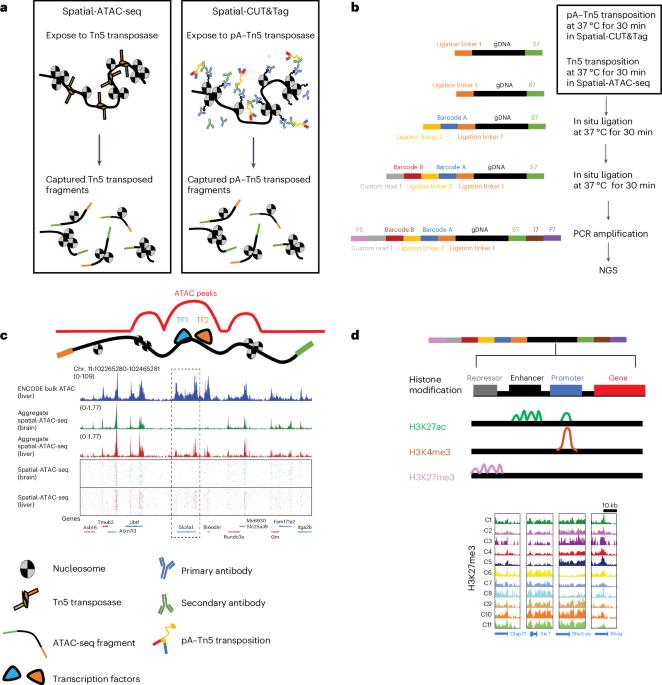

通过组织中的 Tn5 转座和确定性 DNA 条形码进行空间分辨率表观基因组测序。
组织的空间表观遗传图谱可以研究基因调控程序和细胞功能对其局部组织环境的依赖性。在这里,我们概述了两种空间表观遗传组学图谱绘制方法的完整程序:利用靶标和标记(CUT&Tag)化学原位裂解(空间-CUT&Tag)和转座酶可及染色质测序(空间-ATAC-测序)进行染色质可及性的空间解析全基因组组蛋白修饰图谱绘制。这两种检测方法都利用组织内 Tn5 转座来识别基因组 DNA 位点,然后利用微流体确定性条形码来整合空间地址码。此外,这两种方法不需要事先了解特定组织或细胞类型的转录或表观遗传标记,而是允许在 10 μm 像素尺寸水平和单碱基分辨率下进行全基因组逐像素无偏剖析。为支持这些方法的广泛应用,本文将分五个步骤进行详细介绍:(1) 样品制备;(2) 空间-ATAC-测序中的 Tn5 转座或 CUT&Tag 中的抗体控制 pA-Tn5 标记;(3) 文库制备;(4) 下一代测序;(5) 使用我们的定制管道进行数据分析:https://github.com/dyxmvp/Spatial_ATAC-seq 和 https://github.com/dyxmvp/spatial-CUT-Tag。整个过程可在 2-3 天内完成 4 个样本。需要熟悉基本的分子生物学和生物信息学技能,并能使用高性能计算环境。对病理学和标本切片的初步了解,以及组织特定技能中的确定性条形码(如设计多参数条形码面板和创建微流控设备)也是有利条件。在本方案中,我们主要利用空间-ATAC-测序和空间-CUT&Tag 对小鼠胚胎和小鼠大脑的组织区域特异性表观遗传景观进行空间分析,但这些方法也可用于其他物种,无需设计特定物种的探针。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


