Coexpression enhances cross-species integration of single-cell RNA sequencing across diverse plant species
IF 15.8
1区 生物学
Q1 PLANT SCIENCES
引用次数: 0
Abstract
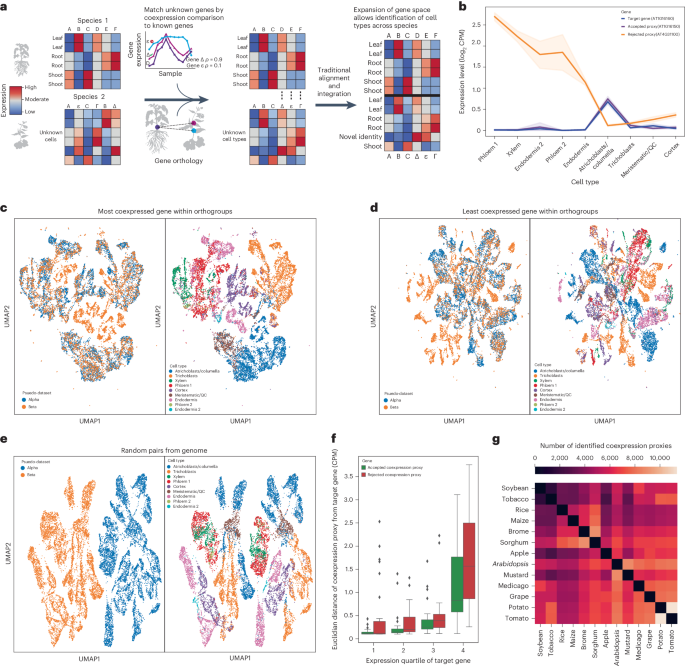
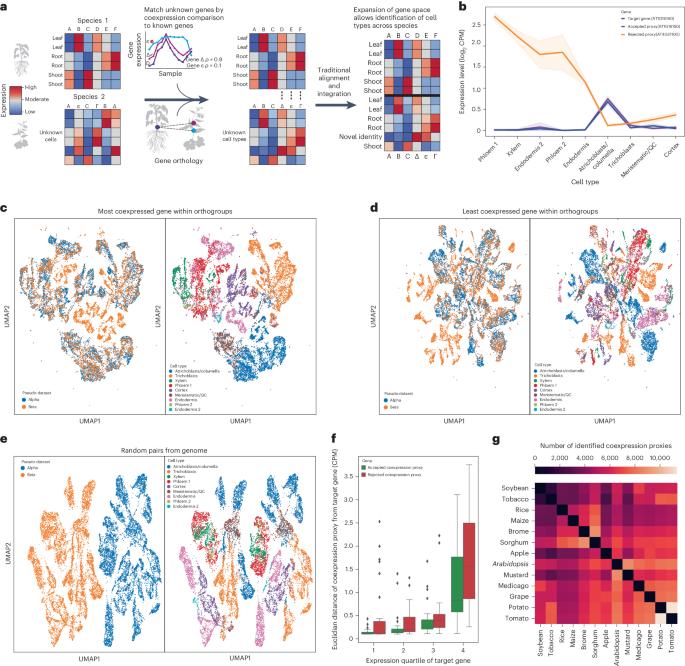
Single-cell RNA sequencing is increasingly used to investigate cross-species differences driven by gene expression and cell-type composition in plants. However, the frequent expansion of plant gene families due to whole-genome duplications makes identification of one-to-one orthologues difficult, complicating integration. Here we demonstrate that coexpression can be used to trim many-to-many orthology families down to identify one-to-one gene pairs with proxy expression profiles, improving the performance of traditional integration methods and reducing barriers to integration across a diverse array of plant species. To enhance cross-species single-cell analysis, the authors find gene pairs with similar expression patterns across 13 species. These coexpression proxies serve as common features in datasets, improving integrative and comparative cell-type analysis.
共表达增强了不同植物物种单细胞 RNA 测序的跨物种整合能力
单细胞 RNA 测序越来越多地用于研究植物基因表达和细胞类型组成所导致的跨物种差异。然而,由于全基因组复制导致植物基因家族频繁扩大,因此很难确定一对一的同源物,从而使整合工作变得复杂。在这里,我们证明了共表达可用于修剪多对多的同源基因家族,以识别具有代理表达谱的一对一基因对,从而提高传统整合方法的性能,减少植物物种多样性整合的障碍。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature Plants
PLANT SCIENCES-
CiteScore
25.30
自引率
2.20%
发文量
196
期刊介绍:
Nature Plants is an online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


