Structural and mechanistic basis for nucleosomal H2AK119 deubiquitination by single-subunit deubiquitinase USP16
IF 12.5
1区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
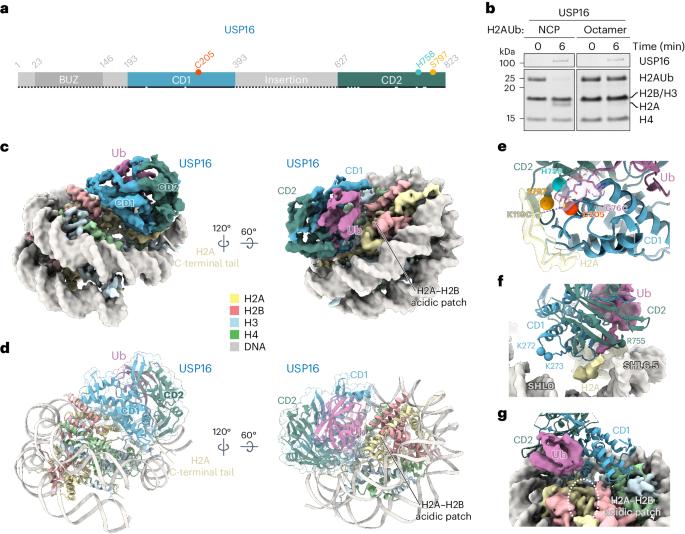
Epigenetic regulators have a crucial effect on gene expression based on their manipulation of histone modifications. Histone H2AK119 monoubiquitination (H2AK119Ub), a well-established hallmark in transcription repression, is dynamically regulated by the opposing activities of Polycomb repressive complex 1 (PRC1) and nucleosome deubiquitinases including the primary human USP16 and Polycomb repressive deubiquitinase (PR-DUB) complex. Recently, the catalytic mechanism for the multi-subunit PR-DUB complex has been described, but how the single-subunit USP16 recognizes the H2AK119Ub nucleosome and cleaves the ubiquitin (Ub) remains unknown. Here we report the cryo-EM structure of USP16–H2AK119Ub nucleosome complex, which unveils a fundamentally distinct mode of H2AK119Ub deubiquitination compared to PR-DUB, encompassing the nucleosome recognition pattern independent of the H2A–H2B acidic patch and the conformational heterogeneity in the Ub motif and the histone H2A C-terminal tail. Our work highlights the mechanism diversity of H2AK119Ub deubiquitination and provides a structural framework for understanding the disease-causing mutations of USP16. The H2AK119Ub is inversely regulated by nucleosomal deubiquitinase. Here the authors report the cryo-EM structure of single-subunit USP16 bound to H2AK119Ub nucleosome, unveiling a fundamentally distinct mode of H2AK119Ub deubiquitination compared to multi-subunit PR-DUB.
单亚基去泛素化酶 USP16 对核糖体 H2AK119 进行去泛素化的结构和机理基础
表观遗传调控因子通过对组蛋白修饰的操纵对基因表达产生至关重要的影响。组蛋白 H2AK119 单泛素化(H2AK119Ub)是转录抑制过程中一个公认的标志,它受到多聚核抑制复合体 1(PRC1)和核小体去泛素化酶(包括主要的人类 USP16 和多聚核抑制去泛素化酶(PR-DUB)复合体)对立活动的动态调控。最近,多亚基 PR-DUB 复合物的催化机理已被描述,但单亚基 USP16 如何识别 H2AK119Ub 核小体并裂解泛素(Ub)仍然未知。在这里,我们报告了 USP16-H2AK119Ub 核小体复合物的冷冻电镜结构,它揭示了一种与 PR-DUB 截然不同的 H2AK119Ub 去泛素化模式,包括独立于 H2A-H2B 酸性补丁的核小体识别模式,以及 Ub 基序和组蛋白 H2A C 端尾的构象异质性。我们的工作凸显了 H2AK119Ub 去泛素化机制的多样性,并为理解 USP16 的致病突变提供了一个结构框架。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Structural & Molecular Biology
BIOCHEMISTRY & MOLECULAR BIOLOGY-BIOPHYSICS
CiteScore
22.00
自引率
1.80%
发文量
160
审稿时长
3-8 weeks
期刊介绍:
Nature Structural & Molecular Biology is a comprehensive platform that combines structural and molecular research. Our journal focuses on exploring the functional and mechanistic aspects of biological processes, emphasizing how molecular components collaborate to achieve a particular function. While structural data can shed light on these insights, our publication does not require them as a prerequisite.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


