A versatile tissue-rolling technique for spatial-omics analyses of the entire murine gastrointestinal tract
IF 13.1
1区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
Tissues are dynamic and complex biological systems composed of specialized cell types that interact with each other for proper biological function. To comprehensively characterize and understand the cell circuitry underlying biological processes within tissues, it is crucial to preserve their spatial information. Here we report a simple mounting technique to maximize the area of the tissue to be analyzed, encompassing the whole length of the murine gastrointestinal (GI) tract, from mouth to rectum. Using this method, analysis of the whole murine GI tract can be performed in a single slide not only by means of histological staining, immunohistochemistry and in situ hybridization but also by multiplexed antibody staining and spatial transcriptomic approaches. We demonstrate the utility of our method in generating a comprehensive gene and protein expression profile of the whole GI tract by combining the versatile tissue-rolling technique with a cutting-edge transcriptomics method (Visium) and two cutting-edge proteomics methods (ChipCytometry and CODEX-PhenoCycler) in a systematic and easy-to-follow step-by-step procedure. The entire process, including tissue rolling, processing and sectioning, can be achieved within 2–3 d for all three methods. For Visium spatial transcriptomics, an additional 2 d are needed, whereas for spatial proteomics assays (ChipCytometry and CODEX-PhenoCycler), another 3–4 d might be considered. The whole process can be accomplished by researchers with skills in performing murine surgery, and standard histological and molecular biology methods. This protocol presents a versatile tissue-rolling technique for spatially profiling the transcriptome and proteome of the whole murine gastrointestinal tract with high spatial resolution.
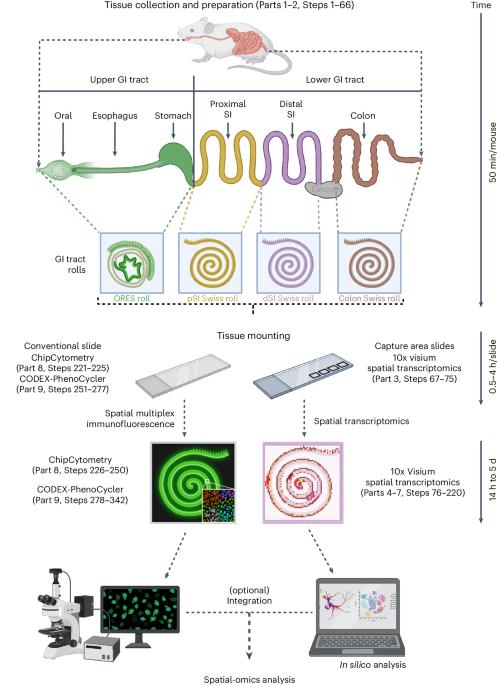
用于整个小鼠胃肠道空间组学分析的多功能组织滚动技术。
组织是一个动态的复杂生物系统,由特化的细胞类型组成,它们相互影响,共同发挥正常的生物功能。要全面描述和了解组织内生物过程的细胞回路,保留其空间信息至关重要。在此,我们报告了一种简单的安装技术,可最大限度地扩大待分析组织的面积,涵盖小鼠胃肠道(GI)的整个长度,从口腔到直肠。利用这种方法,不仅可以通过组织学染色、免疫组化和原位杂交,还可以通过多重抗体染色和空间转录组学方法,在一张载玻片上对整个小鼠胃肠道进行分析。通过将多功能组织滚动技术与最先进的转录组学方法(Visium)和两种最先进的蛋白质组学方法(ChipCytometry 和 CODEX-PhenoCycler)相结合,我们展示了我们的方法在生成整个消化道的全面基因和蛋白质表达谱方面的实用性。这三种方法的整个过程,包括组织卷取、处理和切片,均可在 2-3 d 内完成。对于 Visium 空间转录组学来说,还需要 2 天,而对于空间蛋白质组学检测(ChipCytometry 和 CODEX-PhenoCycler)来说,可能还需要 3-4 天。整个过程可由具备小鼠手术技能以及标准组织学和分子生物学方法的研究人员完成。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Protocols
生物-生化研究方法
CiteScore
29.10
自引率
0.70%
发文量
128
审稿时长
4 months
期刊介绍:
Nature Protocols focuses on publishing protocols used to address significant biological and biomedical science research questions, including methods grounded in physics and chemistry with practical applications to biological problems. The journal caters to a primary audience of research scientists and, as such, exclusively publishes protocols with research applications. Protocols primarily aimed at influencing patient management and treatment decisions are not featured.
The specific techniques covered encompass a wide range, including but not limited to: Biochemistry, Cell biology, Cell culture, Chemical modification, Computational biology, Developmental biology, Epigenomics, Genetic analysis, Genetic modification, Genomics, Imaging, Immunology, Isolation, purification, and separation, Lipidomics, Metabolomics, Microbiology, Model organisms, Nanotechnology, Neuroscience, Nucleic-acid-based molecular biology, Pharmacology, Plant biology, Protein analysis, Proteomics, Spectroscopy, Structural biology, Synthetic chemistry, Tissue culture, Toxicology, and Virology.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


